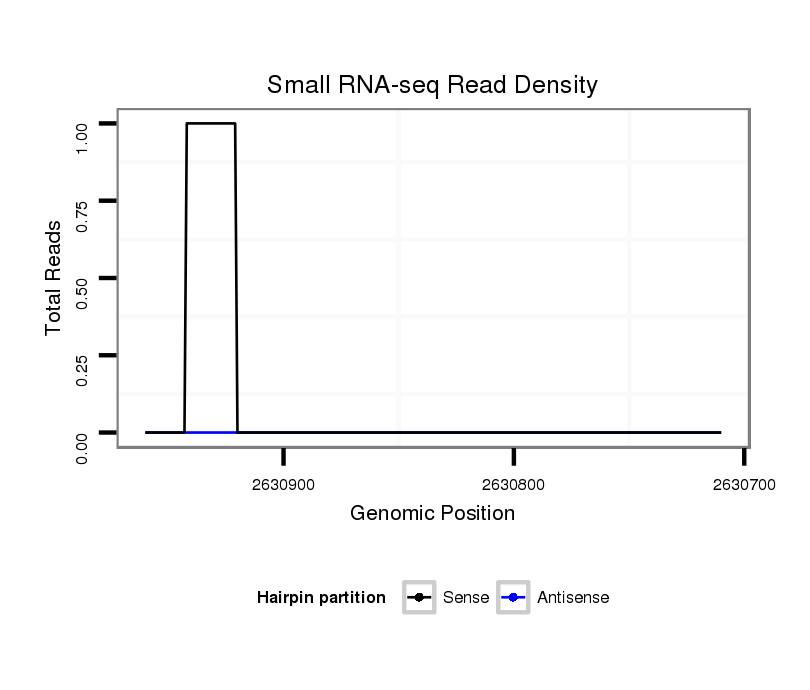
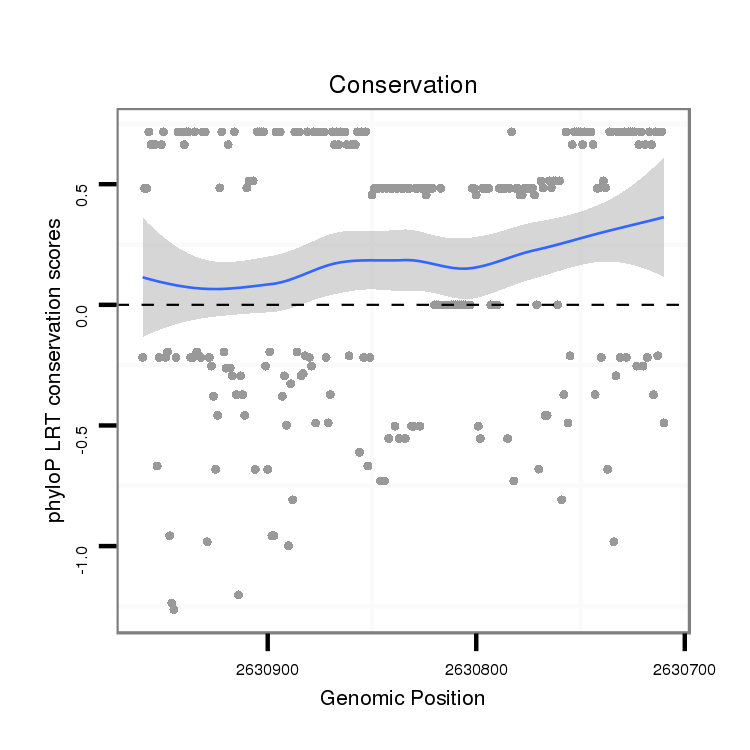
ID:dvi_5021 |
Coordinate:scaffold_12875:2630760-2630910 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_6401:1]; CDS [Dvir\GJ21005-cds]; intron [Dvir\GJ21005-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| -----------------------------#####################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATTTCCCCCGAACTGGGTAACTACCAGACATGATTGGGACACTCTGGCCGGTAAGATATAGGATTATTGACGGTGTCATAATTTTAAAGCGTGTCAATGTCGCACACACAGATAAATAAATGTTTAAATGCATGTACTATATCAATATATTATGTTTTATGTTTTATATATATATTTTTTTCCTAATTGCTTGGTACTTAAGCTGACTAATCTTATCGAGCTGCAACGATAAATAATACTTGTACGATTTC **************************************************((((((((((((((((((((.((((.(((.((((..((((((.....((((.........)))).....))))))))))..)))))))...))))))......))))))))))))))..............((.........)).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060667 160_females_carcasses_total |
V047 embryo |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|
| ..................AACTACCAGACATGATTGGGAC................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................................................................................TCATGTACAATATCAATATACT.................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ......................ACCCGGCTTGATTGGGACAC................................................................................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ...........................................................AGCATTATTGACGATGTC.............................................................................................................................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 |
|
TAAAGGGGGCTTGACCCATTGATGGTCTGTACTAACCCTGTGAGACCGGCCATTCTATATCCTAATAACTGCCACAGTATTAAAATTTCGCACAGTTACAGCGTGTGTGTCTATTTATTTACAAATTTACGTACATGATATAGTTATATAATACAAAATACAAAATATATATATAAAAAAAGGATTAACGAACCATGAATTCGACTGATTAGAATAGCTCGACGTTGCTATTTATTATGAACATGCTAAAG
**************************************************((((((((((((((((((((.((((.(((.((((..((((((.....((((.........)))).....))))))))))..)))))))...))))))......))))))))))))))..............((.........)).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060666 160_males_carcasses_total |
M047 female body |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................GAAAAGGAGTAACGAATCATGAA.................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................TAATAACTGCCAAAGTAAT.......................................................................................................................................................................... | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 |
| ..AAGGGGGCGTGAACCATTA...................................................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 |
| ....................................................................CTGCCACCGTATTGAAATA.................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:2630710-2630960 - | dvi_5021 | ATTTCCCC-CGAACTG---GGTAACTAC-CAGACATGATT-------------GGGACACTCTGGCCGGTA-AGATATAGGATTATTGACGGTGTCATAATTTTAAAGCGTGTCAATGTCGCACACACAGATAAATAAATGTTTAAATGCATGTA----CTATATCAATATATTATGTTTTATGTTTTATATATATATTTTTTTCCTAATTGCTTGGTACTTAAGCTGACTAATCTTATCGAGCTGCAA------CGATAAAT-AATACTT-GTACGATTTC |
| droMoj3 | scaffold_6496:22601957-22602176 + | ATTTCCCA-TGAACCCTTTCATAACTATGTAGATATAACTAA--------------ACTCACTAACT-------ATATG-GGCTATTGCATCTGTCGTAACTGTAAAACATGTCAATGTCGCAGATAAAGATACAGATATATATATATATATATAAATTCTAT---A--------------ATGCATTAT----ATATATTGTTCCTAATTG------------CTTGACTAATCTTATCAA-T---AATGTTTTTGATAAAT-AATGCTC-GTACAATTTC | |
| droGri2 | scaffold_15112:4754355-4754543 - | T--TCCCATCGATTAAGGTTGTAACTACGCAAGCATTTTGTAACTGAATGATATAGATACTATACATAGTATATATATATATATATGTATGCTATAGCATTTGTAAAGGATGTCAATGCCGCACACAAT-----------------------------------------------------------------------T------------GTGACACTT-ACTTCGCTAATCTTATCA-GCTGAAA------ATAAAATTAAATACTTGGAACAACTTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 04:55 AM