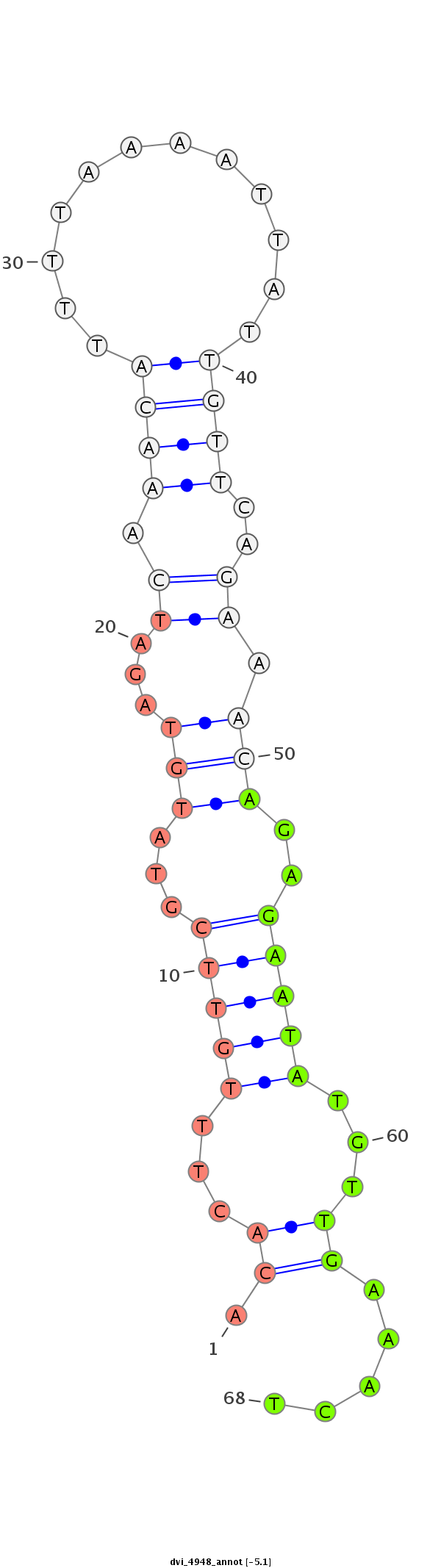
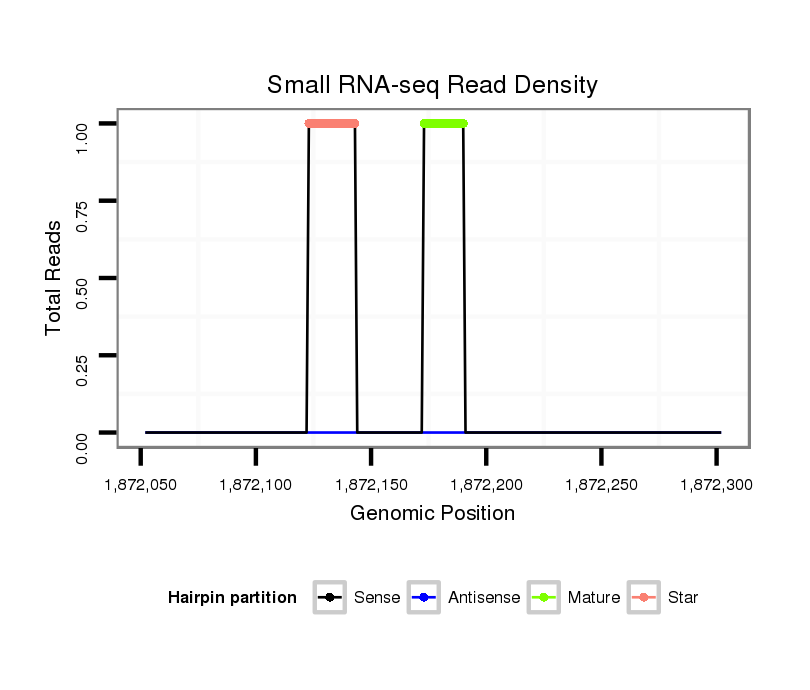

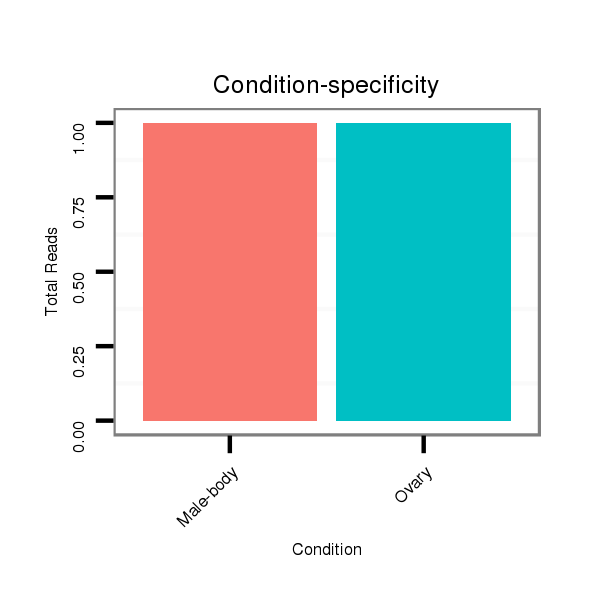

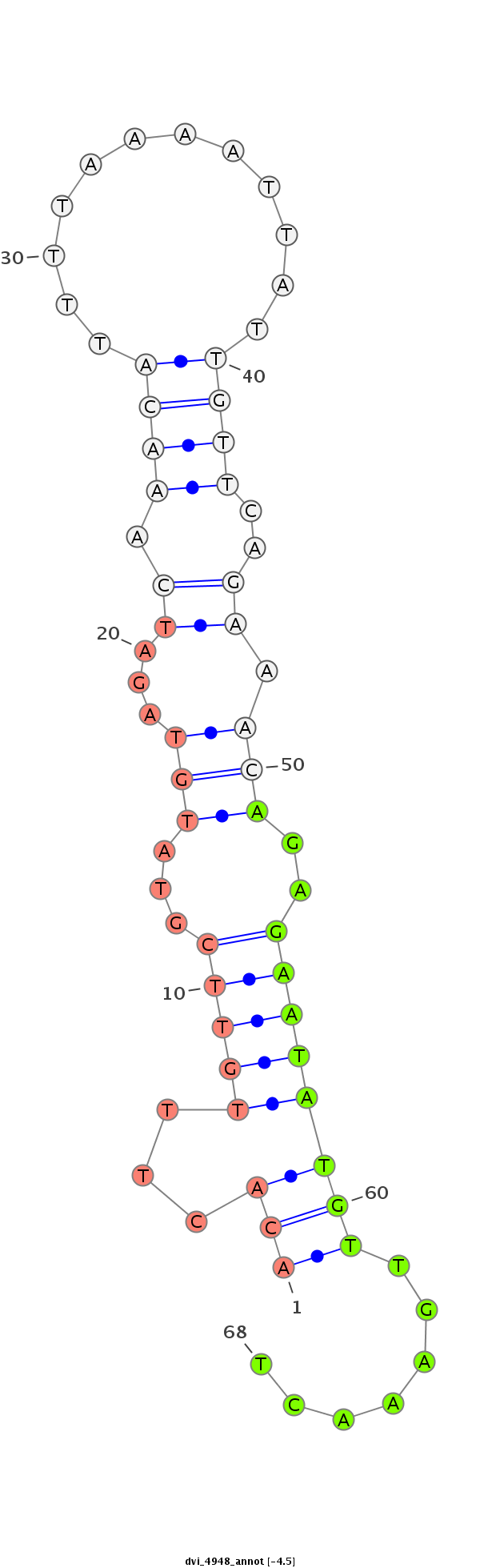
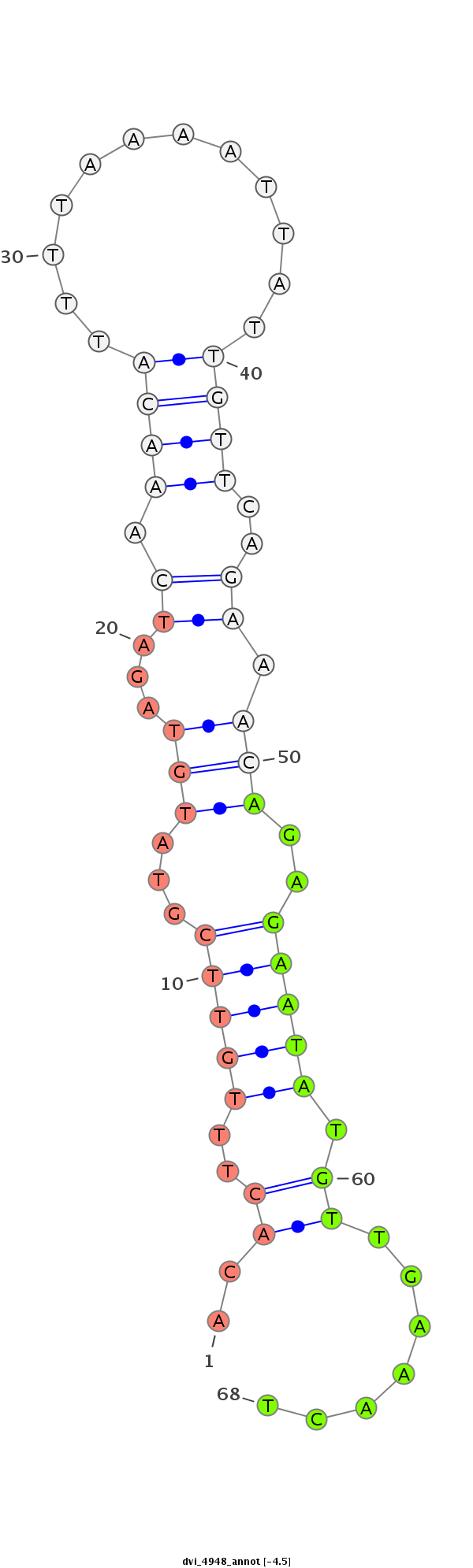
ID:dvi_4948 |
Coordinate:scaffold_12875:1872102-1872252 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -4.8 | -4.5 | -4.5 |
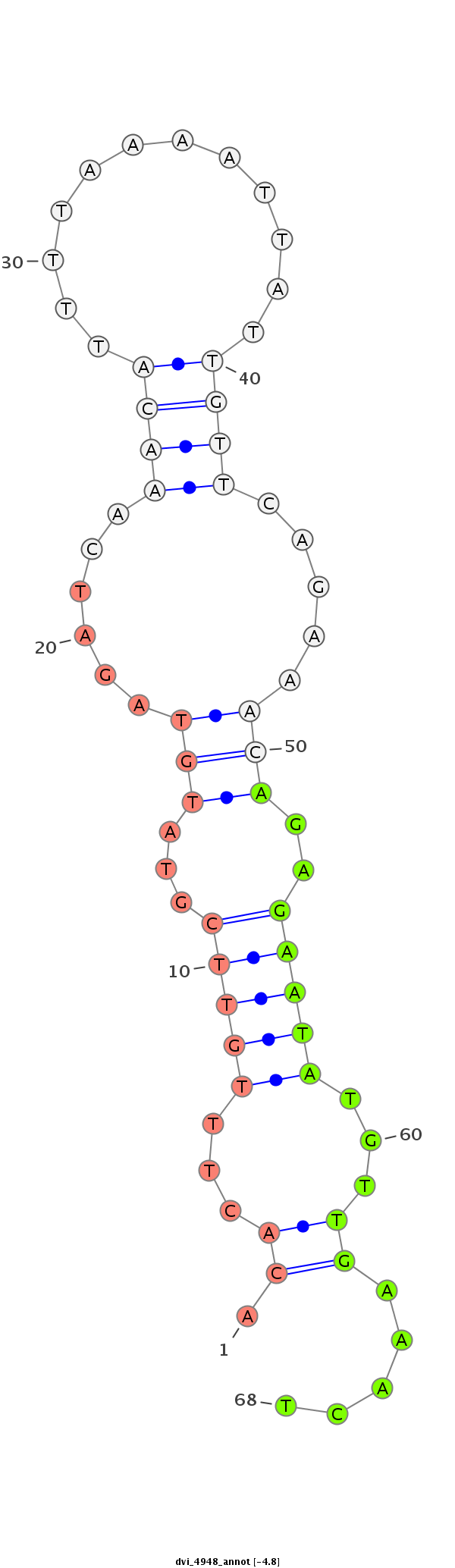 |
 |
 |
exon [dvir_GLEANR_6685:2]; CDS [Dvir\GJ21318-cds]; intron [Dvir\GJ21318-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGTATCGATAGTTTCGCCGCTGCGCAACAATTGTCAATCGTTTATCAGCTGTTATTATAAATTTAAGCAAGACACTTTGTTCGTATGTAGATCAAACATTTTAAAATTATTGTTCAGAAACAGAGAATATGTTGAAACTGTAAGTAGATTGCTAGACCAATGCCGATCCGTTCCAGACGCTGATAATACCATTTTCATCAGCTGCCGCCTAGAAGAATGCATGCAGCTGCGCAGCTCCCAATGGTGGCCTC ***********************************************************************.((...(((((...(((...((.((((............))))..)).)))..)))))...)).....**************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060660 Argentina_ovaries_total |
V116 male body |
M028 head |
SRR060668 160x9_males_carcasses_total |
V047 embryo |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................AGAGAATATGTTGAAACT................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................ACACTTTGTTCGTATGTAGAT............................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TATTGTTCAGAAACAGCGAA............................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TTCCAGACTCTGATCATTCC............................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................GCTGCTAGTATAAATTTAAGT....................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................GAAGCTTTGCTCGTATGTAG................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| AGAATCGATACTTTGGCCGC....................................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................AACAGTTGTCAATCGTTGTTC............................................................................................................................................................................................................. | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TCATAGCTATCAAAGCGGCGACGCGTTGTTAACAGTTAGCAAATAGTCGACAATAATATTTAAATTCGTTCTGTGAAACAAGCATACATCTAGTTTGTAAAATTTTAATAACAAGTCTTTGTCTCTTATACAACTTTGACATTCATCTAACGATCTGGTTACGGCTAGGCAAGGTCTGCGACTATTATGGTAAAAGTAGTCGACGGCGGATCTTCTTACGTACGTCGACGCGTCGAGGGTTACCACCGGAG
****************************************************************************************************************.((...(((((...(((...((.((((............))))..)).)))..)))))...)).....*********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
V053 head |
SRR060663 160_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
M061 embryo |
SRR060689 160x9_testes_total |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TACGGCTCGGAAGGGTCTG......................................................................... | 19 | 3 | 7 | 4.57 | 32 | 12 | 10 | 1 | 0 | 3 | 0 | 2 | 2 | 0 | 1 | 1 | 0 |
| .CCTTGTTATCAAAGCGGCGA...................................................................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GTAAAGGTAGTCGTCGGCTGAT........................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CGATCTGGTTGCTGCTAGGG................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................ACAGTCGGCAATAATATTTATA........................................................................................................................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TGGTAAAAGTAGTACACG.............................................. | 18 | 2 | 15 | 0.13 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................CTAAGAAATAGTCGACAATA.................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:1872052-1872302 + | dvi_4948 | AGT--------ATCGA--TA-G-TTTCGCCGCTGCGCAACAATTGTCAATCGTTTATCAGCTGTTATTATAAATTTAAGCAAGACACTTTGTTCGTATGTAGATCAAA--CATTTTAA------AA--------TTATTGTTCAGA--------AACAGAGAATATGTTGAAACTGTAAGTAG-ATTGCTAGACCAATG----------CCGATCC----GTTCCAGA--C-GCTGATAATACC--ATTTTCATCAGCTGCCGCCTA--GAAGAATGCA-TGCA------------------------------------GCT--------------GCGCAGCTCCCAATGGTGGCCTC |
| droMoj3 | scaffold_6496:13618480-13618741 - | ATCGGTTTTATTATAACAAAATATTT-ACCGATAT-CAAT-AGCTTCAATAGGGAATCAGCTGTTGCTGAAAATCAAAACAAA-AACTTTAGACAAA--------------ATTTTAAATTTTTAA--------TTGTTGTCAGTC--------ACTTAAAAATATGTTAAAACTGTAAGTACGGAAGTGCGTTTAACACATATTTATTATATTTA----T---GAATACC-GCTGACATT-CC--CTTTGTAGTAGCTGTCGTTTA--GAAGAATGCAAAAGA------------------------------------GCC--------------G-TCAATTCGCAACAATTTCGTC | |
| droGri2 | scaffold_15112:3974533-3974775 + | -GT--------ATCGA--TA-A-CATTG----------CT-ATCGTCAATCGTCAGTCAGCTGTTTGTTGATAC--AAACACAACATTTTGTATGGATGTAAATAATAGTAATATTTA------AA--------TTTTTGTACACAACTTGAA-----GAAAATATGTTGAAAATGTAAGTAG-TTT--TATATCCATG----------GAAATCA----T---CAATAACTCATGCAATTATC--CCGCCTATCAGTTGTCGCCTC--GACGAATGCAGCAGACAAAT------------------------------------------------TCGAACTTCCCAATGGCTGCGAC | |
| droWil2 | scf2_1100000004558:483611-483795 - | AAT--------TTTAGTAAAATATTTAGTTA-------AT-TTT------------------------------------------------------GTAAAACAAA--GAAATC-A------AATTTATGTGA---TATTGGCC--------GACCAATAATATGTTGAGTTTGTAAGCATGTCAACG-------------------GTAACCATTGGCTTTCATTTACCCATACTTTAATCTTC-C------------TCTATT--AGCCGGATTTGTAAAGT---------------------------------------------------ACAACGCTTCCAATCCTGTCAAA | |
| dp5 | 3:2746139-2746379 - | TGT--------ATTGACATG-G-TCTGGCAA-------CC-TTCGTCAATC--CAGTCAATTGTTGATTGTTGTTATTGTTAA-AAATTCATTTAGA--------------TATTTAA------AATTTGTAAAA---AA-------TTGGTACGAACAGAAACATGTTGAGTCAGTAAGTAAGATAGCTACCCTGCCAG----------------TTGGCCA----------ATAATAATACCTCA-G------------TGTCTT--CGCAGTTACAGCAAAGAGGTGCTCCTCAG---------------A----------CACTGG--TCCTGGCGGAACTTTCAGTCCTGCCGAC | |
| droPer2 | scaffold_2:2936148-2936387 - | TGT--------ATTGA-ATG-G-TCTGGCAG-------CC-TTCGTCAATC--CAGTCAATTGTTGATTGTTGTTATTGTTAA-AAATTCATTTAGA--------------TATTTAA------AATTTGTAAAA---AA-------TTGGGACGAACAGAAACATGTTGAGTCAGTAAGTAAGATAGCTACCCTGCCAG----------------TTGGCCA----------ATAATAATACCTCA-G------------TGTCTT--CGCAGTTACAGCAAAGAGGTGCTCCTCAG---------------A----------CACTGG--TCCTGGCGGAACTTTCAGTCCTGCCGAC | |
| droKik1 | scf7180000302385:177687-177872 - | TGT--------TTTTG---------------------------------------------------------------------------------TTTACATGAAC--CAATTCAA------AATTCTTAGAA---AT-------TTG----------GGCTATGCTTATTCTGTGAGTAT-ACCGCAGC---TTCAG----------------ATAGTCA----------GG-ATCTCATCCCC-A------------TATCCTTACACACTTACAGCAAAGAAACGTTCCGGATCTTTGGGAGCGCGCGGCTTGAATCCGCGCGGATCTGCCGGCGGA-------------GCTTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/17/2015 at 04:26 AM