
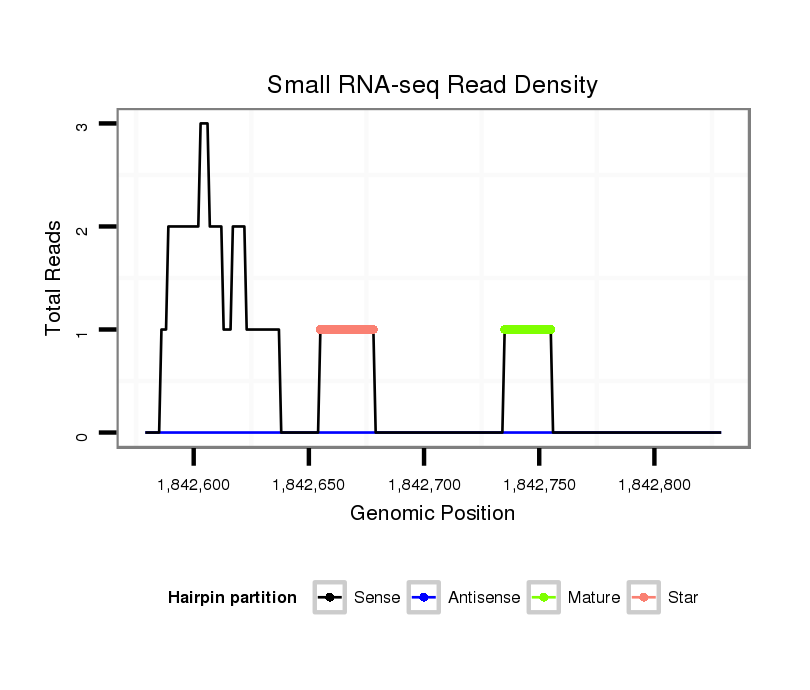
ID:dvi_4935 |
Coordinate:scaffold_12875:1842629-1842779 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -21.3 | -21.2 | -21.1 |
 |
 |
 |
CDS [Dvir\GJ21314-cds]; exon [dvir_GLEANR_6681:1]; intron [Dvir\GJ21314-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CCGGCGAAAGTTCCTGGAAAGAACGGGCCTGAGGAAGCGCAAATGTGAAGGTGTATATCTATCTATTTTATTATTATTACCAACAGTTCAGGTATACTTTAAATGGCACTCCCTTGAACAAGTTACCTCCTTGTTCAGTTGGCCGAAAGTCTGAGTCACTTGGTCAACTTACATCTACTGAGTACTGCAGTTGTATTAGTATTATTTGTTATATATTTTTGTTTATCTTTTTTGTAATCTTATCGTTAAGC ****************************************************************************..(((((...(((((....(((((...((((......(((((((((.......)))))))))...))))))))))))))....))))).............************************************************************************** |
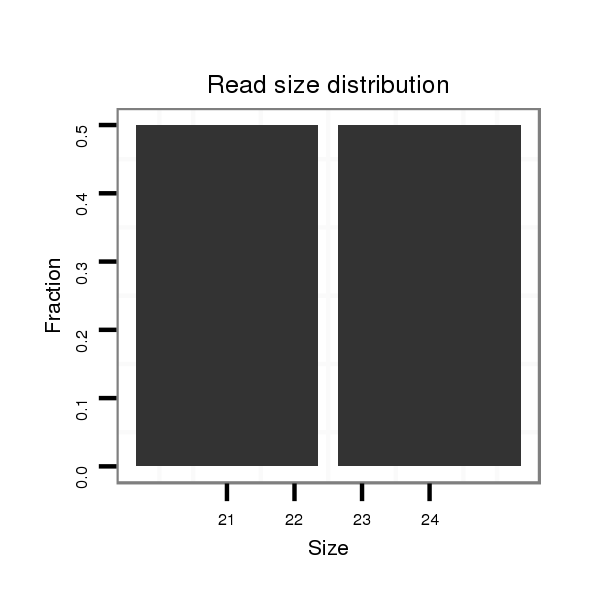
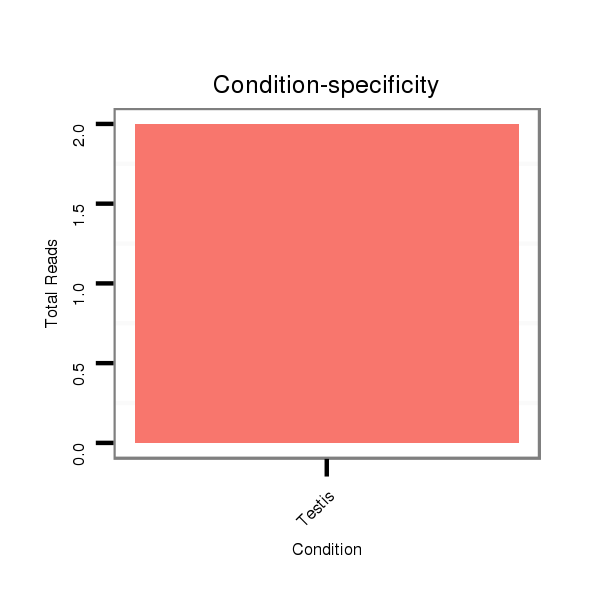
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060678 9x140_testes_total |
SRR060683 160_testes_total |
V053 head |
SRR060663 160_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR1106729 mixed whole adult body |
SRR060660 Argentina_ovaries_total |
SRR060654 160x9_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060673 9_ovaries_total |
V047 embryo |
M047 female body |
V116 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR1106723 embryo_12-14h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................GGGCCTGAGGAAGCGCAAAT............................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................GCAAATGTGAAGGTGTATATC................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......AAGTTCCTGGAAAGAACGGGC............................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TTCCTGGAAAGAACGGGCCTGAGG......................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................CACTTGGTCAACTTACATCTA.......................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TTACCAACAGTTCAGGTATACTTT....................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCTTGTTGAGTTGGCCGAGA....................................................................................................... | 21 | 3 | 5 | 0.80 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................AACGGGCCTAAGGAAGCGAA.................................................................................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTGGGCATCTTACATCTGCTG....................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................AACGGTCCTAAGGTAGCGCA.................................................................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AAGAACGGGCGTGAG.......................................................................................................................................................................................................................... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................AACGGGCCTAAGGGAGCGAA.................................................................................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................ATTGTTTATCTTTTTTGTCAT............. | 21 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTTTGATTATCTTTTTTGT................ | 19 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ............................................................................................................ATCACCTGAACAAGTTACCT........................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ATAACGGTCCTGAGGTAGC..................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................AACGGTCCTAAGGAAGCGAA.................................................................................................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTGGTCATCTTACATGTTCT........................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................ACGGTCCTAAGGAAGCGAA.................................................................................................................................................................................................................. | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................ACGGGCCTAAGGTAGCGAA.................................................................................................................................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................CACTTGGACAACTTACAG............................................................................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................TTCAGGTATTCGGTAAATG.................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GGCCTAAGGTAGCGCAATT............................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GGCCGCTTTCAAGGACCTTTCTTGCCCGGACTCCTTCGCGTTTACACTTCCACATATAGATAGATAAAATAATAATAATGGTTGTCAAGTCCATATGAAATTTACCGTGAGGGAACTTGTTCAATGGAGGAACAAGTCAACCGGCTTTCAGACTCAGTGAACCAGTTGAATGTAGATGACTCATGACGTCAACATAATCATAATAAACAATATATAAAAACAAATAGAAAAAACATTAGAATAGCAATTCG
**************************************************************************..(((((...(((((....(((((...((((......(((((((((.......)))))))))...))))))))))))))....))))).............**************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M028 head |
SRR060679 140x9_testes_total |
M027 male body |
SRR060681 Argx9_testes_total |
V047 embryo |
SRR060666 160_males_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................CTCGTGACGTCAACATAAC..................................................... | 19 | 2 | 3 | 1.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TCGTGACGTCAACATAA...................................................... | 17 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................AGAGATAGATAAAATAGTAGTA.............................................................................................................................................................................. | 22 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................AAAAAACCTTAGGATAGC...... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................AGTCATCCGGCTCTAAGACT................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................AGTGAACCAGTTGAAGGAAAA........................................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................GTGAACCAGTTGAAGGAA............................................................................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
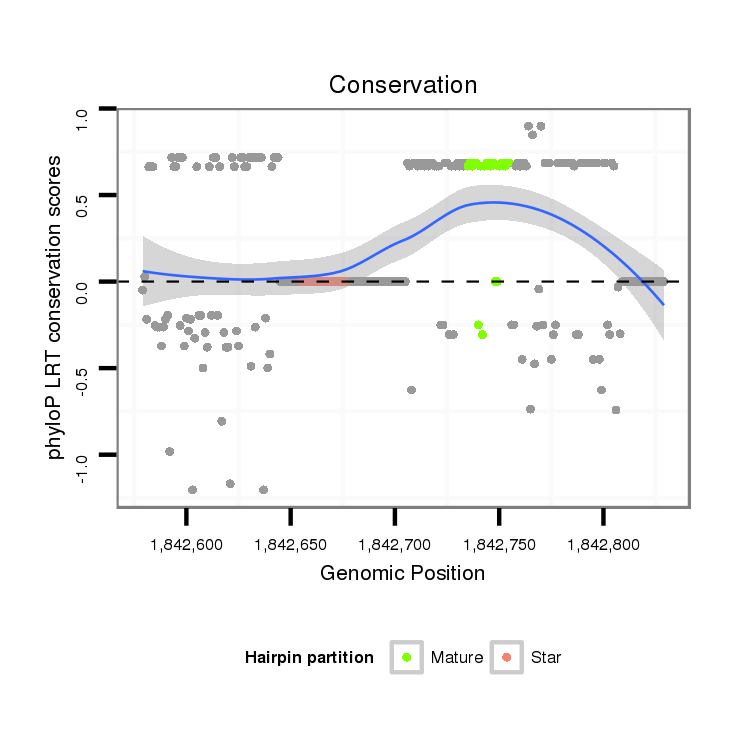
| droVir3 | scaffold_12875:1842579-1842829 + | dvi_4935 | CCGGCGAAAGTTCCTGGAAAGAACGGGCCTGAGGAAGCGCAAATGTGAAGGTGTATATCT------A----TCTATTTTATTATTATTACCAACAGTTCAGGTATACTTTAAATGGCACTCCCTTGAACAAGTTACCTCCTTGTTCAGTTGGCCGAAAGTCTGAGTCACTTGGTCAACTTACATCTACTGAGTACTGCAGTTGTATTAGTAT-TATTTGTTATATATTTTTGTTTATCTTTTTTGTAATCTTATCGTTAAGC |
| droMoj3 | scaffold_6496:13650696-13650775 - | CAAGCGGTTAATCTTGGAGAAAGTATGCCGGAGGAAGCCCAAGTGCAAAGGTGTTTATTT------CTTTAACTAT-----------------------------------------------------------------------------------------------------------------------TCCTGTT------------------------------------ATC--------------------- | |
| droGri2 | scaffold_15112:3943596-3943671 + | GCGGCGAAAATATATGGAAAAGGCCGGTTTTCGAAAACCACCCTGCAAAGGTCTATATGCACTTTTATCTAACTAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droWil2 | scf2_1100000004954:2741781-2741884 - | CC---------------------------------------------------------------------------------------------------------------------------------------TCGTTGTTCAGTTGGCTAAATGACTGAGTCACTTAGACAACT--CATCTATCGAGGACTGCTCATATATGTATATATATTTGAAATATATGTTGCTTCTTCCCT--------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/17/2015 at 04:16 AM