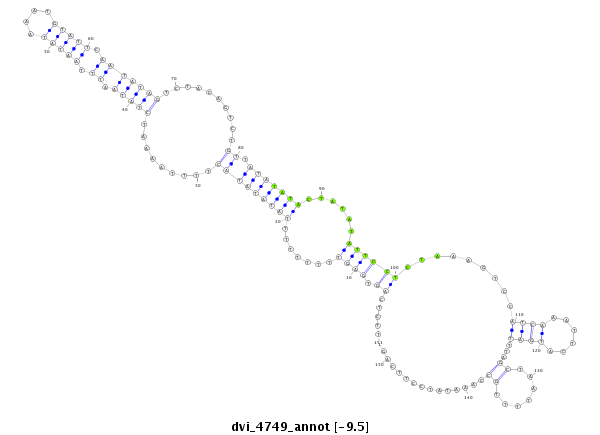
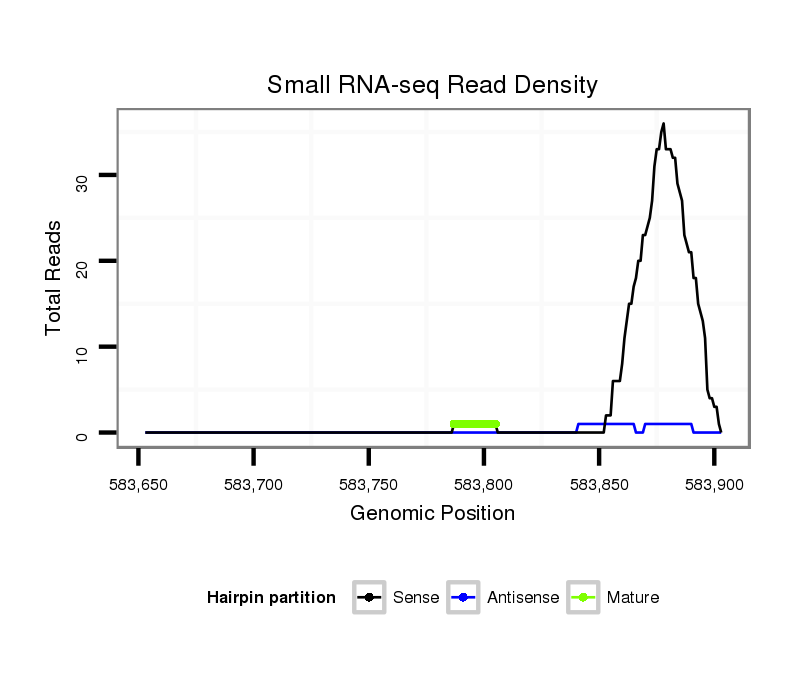
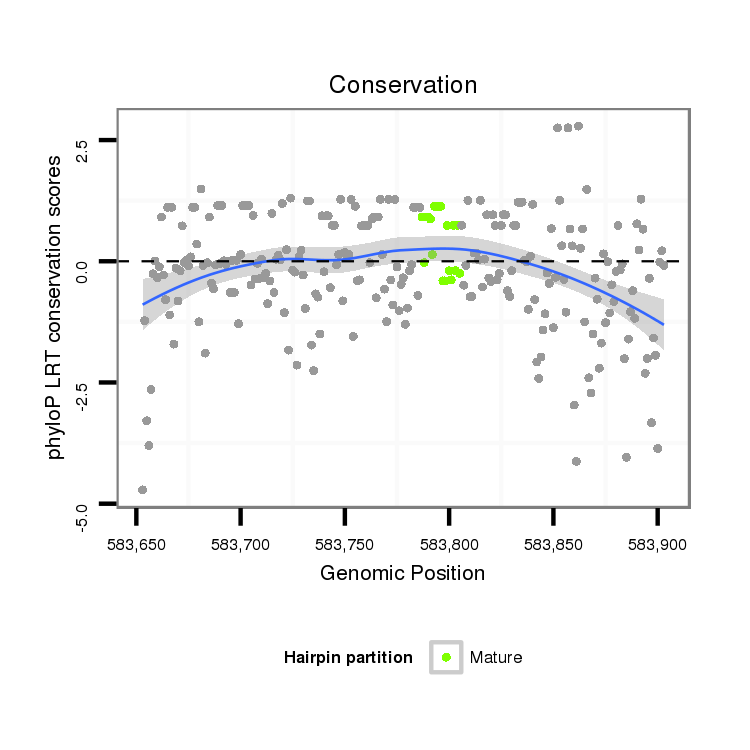
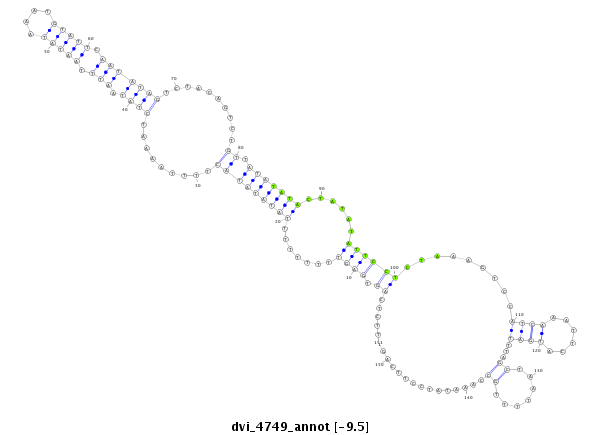
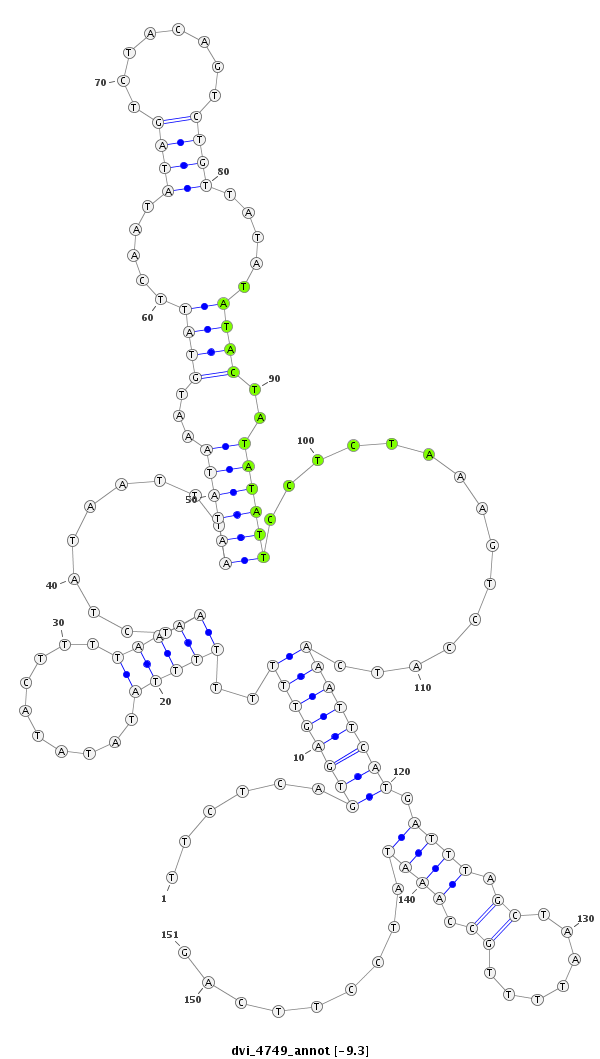
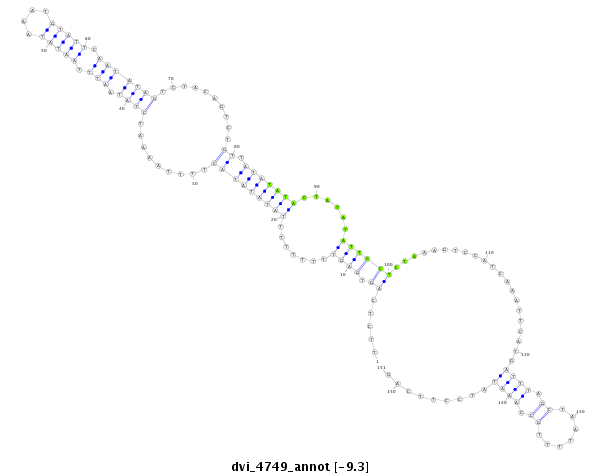
| CCCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACCTTTCAGAACACAATTGGCTTCACTTCAG-----CCACGGGGCCATTCCCCAGGCAGCC | Size | Hit Count | Total Norm | Total | M044
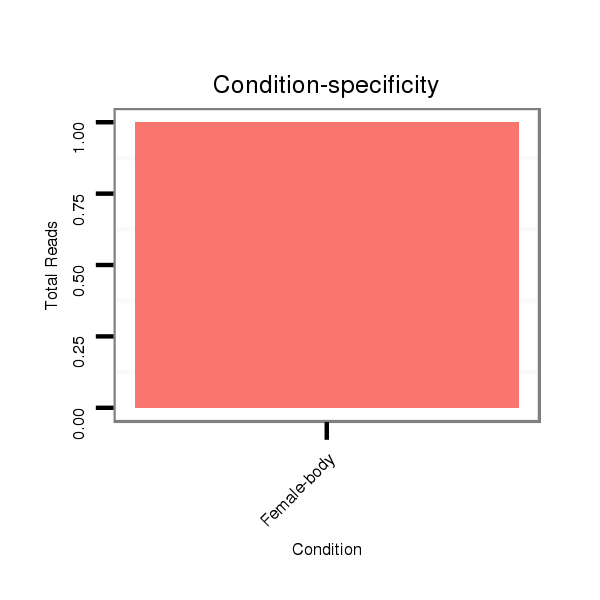
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| .......................................................................................................................................................................................................................................................................................................................................................................................CTTCACTTCAG-----CCACGGGGCC............... | 21 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................GCTTCACTTCAG-----CCACGGGGC................ | 21 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................CACTTCAG-----CCACGGGGCCATT............ | 21 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................TTCACTTCAG-----CCACGGGGCCATTCCCC........ | 27 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................GAACACAATTGGCTTCACTTC................................ | 21 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................ACACAATTGGCTTCACTTCAG-----CCACGG................... | 27 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................ATTGGCTTCACTTCAG-----CCACGGG.................. | 23 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................CACGGGGCCATTCCCCAGGCAGCC | 24 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................GGCTTCACTTCAG-----CCACGGGG................. | 21 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................TTCACTTCAG-----CCACGGGGCCA.............. | 21 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................AATTGGCTTCACTTCAG-----CCACGGGGCC............... | 27 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................TTCACTTCAG-----CCACGGGGCC............... | 20 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................CTTCAG-----CCACGGGGCCATTCC.......... | 21 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................ATTGGCTTCACTTCAG-----CCACGGGGC................ | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................CAATTGGCTTCACTTCAG-----C........................ | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................CTTCACTTCAG-----CCACGGGGCCA.............. | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................CACTTCAG-----CCACGGGGCCA.............. | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................ACAATTGGCTTCACTTCAG-----C........................ | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................AACACAATTGGCTTCACTTCA............................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................CACTTCAG-----CCACGGGGCCATTCCCCAGGC.... | 29 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................CTTCACTTCAG-----CCACGGGGCCT.............. | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................TCAG-----CCACGGGGCCATTCCCC........ | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................GGCCATTCCCCAGGCAGC. | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................CACTTCAG-----CCACGGGGCCATTCCCC........ | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................TCACTTCAG-----CCACGGGGCCA.............. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................CTTCACTTCAG-----CCACGGGGCCATTCC.......... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................TTCACTTCAG-----CCACGGGGT................ | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................CCACGGGGCCATTCCCCAGGCAGCC | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................TCACTTCAG-----CCACGGGGC................ | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................GAACACAATTGGCTTCACTTCAG-----CCAC..................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................CACTTCAG-----CCACGGGGCCAT............. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................GCTTCACTTCAG-----CCACGGGG................. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................TCACTTCAG-----CCACGGGGCC............... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................CTTCACTTCAG-----CCACGGGGCCATTCCCCA....... | 29 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................ACACAATTGGCTTCACTTC................................ | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................AACACAATTGGCTTCACTTCAG-----C........................ | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................CACAATTGGCTTCACTTCAG-----......................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................ACACAATTGGCTTCACTTCAG-----C........................ | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................CTTCACTTCAG-----CCACGGGGC................ | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |