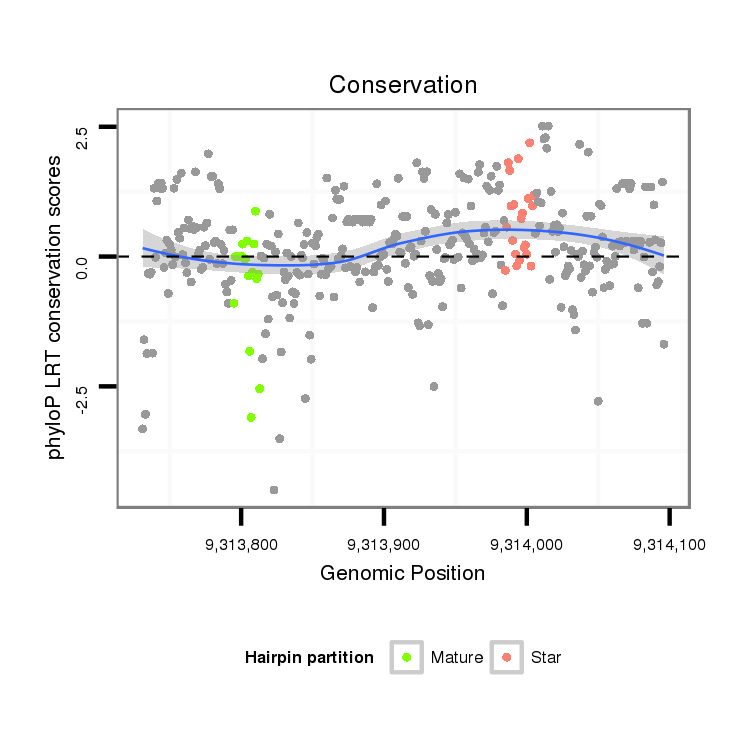
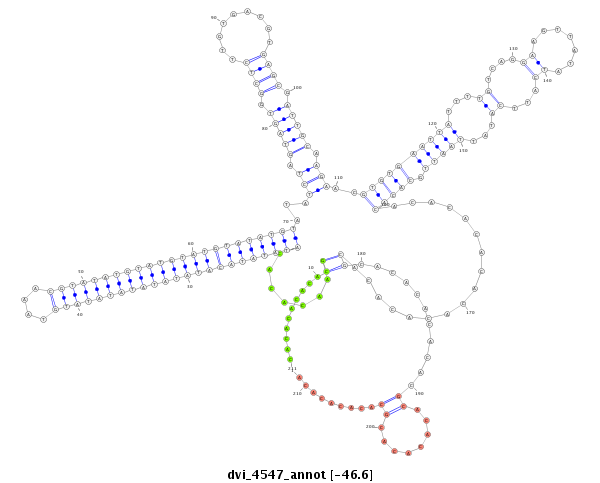
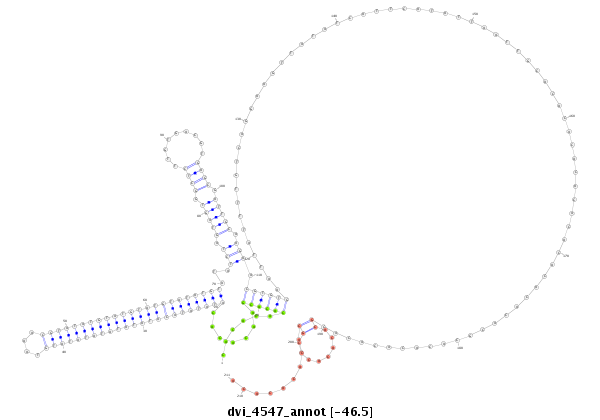
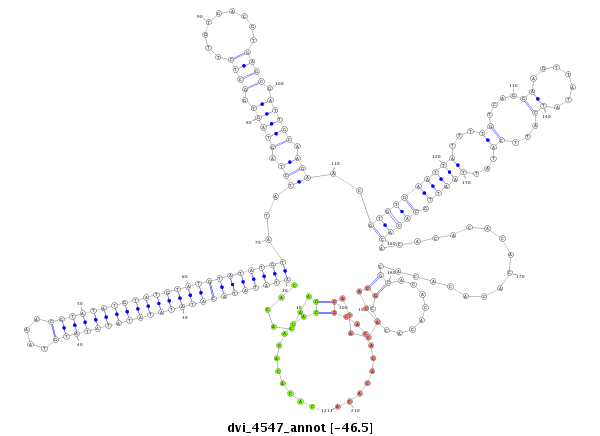
| droVir3 |
scaffold_12855:9313731-9314096 + |
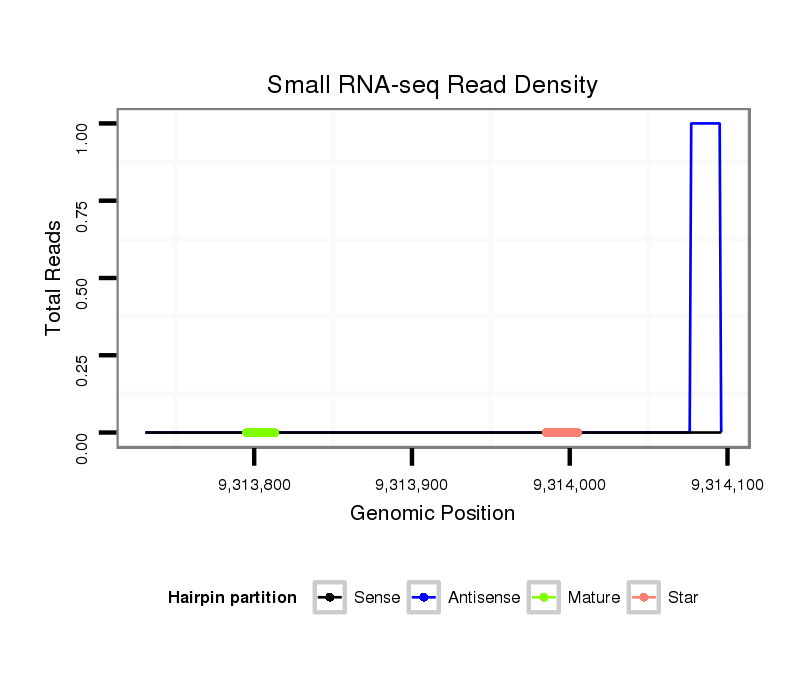
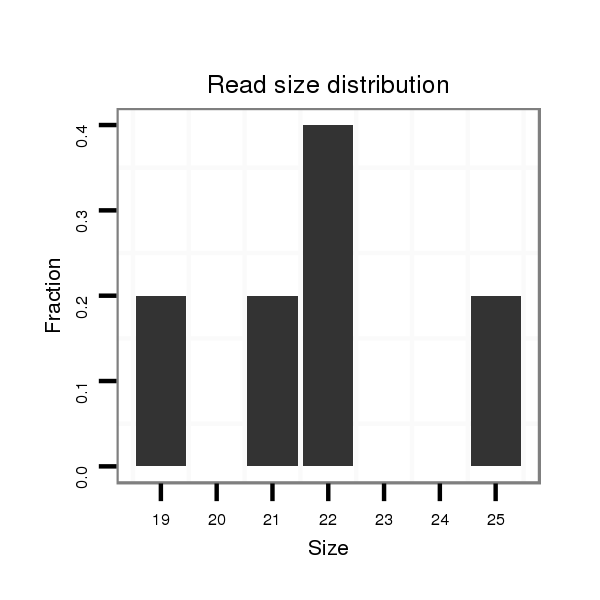
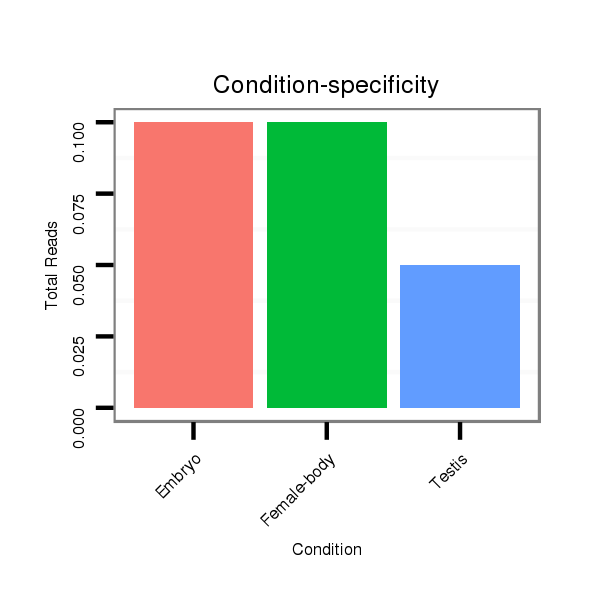
dvi_4547 |
CCAGCCAGCGAACAGTGTCTCC-GT--------------------------CTCCGTTGACCTGCACGTACTCAGCGGTGAGTCCTTGACACACACACACACGAACACACATATATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATATATAT---------------------ATATG------------------TAAACGT--------------ATATGTATGTATGTATATGTA-T----------------------ATCTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAGTGGCTCTTGTGACGTGAGCGATTGCAAGAACGTGTGAATTATTTTGTCAG--GAAGTTATATCATTCAT------------------ATTAATTG-------------C--ACACACACAC-----------------------------------------------------------ACA-CACACAC----ACGCACA--CACA--------------------------------------------------------------------------------------------------C------ACGCACACACACGCACACACA------CATACACACA------------------------CACACACACACA------------------------------CACATT----------------------------------------------------CA---------------TTATTGCCAACAGGTGAATACT----CC------------GCTTCGGCTATCATGTCCGGCGCAGCAGAGTGTCAACAG |
| droMoj3 |
scaffold_6540:24402465-24402811 + |
dmo_2048 |
CCAGCCATCGAACAGCGAGTCC-GT--------------------------CTCCGTTGACTTGCACGTGCTCAGCGGTGAGTCCT-----------------------------------------------CCCTCAATATTTTTTCGTCAACATATTGATAGTGAAGA-GTGTAC----------------------------------------------------------------------------------------------------------------------------ATACACATACATG---TATATGTAT----------------------------------------------------------------GTATGTATAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAGTGGCTCTTGTGACGTGAACGATTGCGAGTGCGTGTGAATTATTTTGTCAG--CAAGTTAAATGATTCAT------------------ATTAATTGCAACTGTCA----CACT----CACAC-----------------------------------------------------------TCA-CACACAC----TC--------------------------------------------------------------------------------------------------------------------------ACACA------CACT------CACTCACAC--------------------------ACACGCACACG----------------------------CAC----------ATC------------TCT-----------------------------CT---------------CTTTTGCCAACAGGTGAATACT----CG------------GCTTCGGCTATCATGTCCGGCGCTGCTGAGCGCAAACAG |
| droGri2 |
scaffold_14830:1778970-1779354 + |
|
CCAGCCATCAAACAGCGTTTCT-GT--------------------------CTCCGTCGACCTGCATGTCCTCAGCGGTAAGTCCAGCATAG----C--------------------------------------------------------------------------TATGCACACA--------TACATACA----------------------------------------------------------TACTGACATACATACAT----------------------------AGATT---------------------------------------------------------CACACAT--------------AGATACATA-------------CA----------------------------------------------------------------------------------------------------TTTATATTTAC------------------------------------------------AA-------------------------------------------------------------------------------------------------------------------------------------T---------------AAACAAGCATTATTTTATGTTGTAACTACTCTGAAGTGATAAATGGCGAGTGATTATGTTTTTTCTTTTTATATCAAGCTTAATGATTCAT------------------ATTAATTGCAACTGTCG----C----------------------------------------------------AACACACTCACACACACACAC-----------------AAA--CA----------------------------------------------------------------------------------------------------------------------CACACATACG------CACACGCACA------------------------CATACAAACACA------------------------------CAC--TTATACATT------------ACT-----------------------------TA---------------CTATTCCCAACAGGTGAATATT----CC------------GCATCGGCCATCATGTCCGGAGTGGCCGAGCGTACGAAG |
| droWil2 |
scf2_1100000004902:428848-429103 - |
|
CCAGCCATCAAACAGTATCTCT-GT--------------------------CTCCGTCGACCTGCACGTCCTCAGCGGTGAGTTGG-----------------------------------------------CCC------------------------------------ACAGACCTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T----------------------ATCTAGCTTTTGTTTTTATGCCTGTGTCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTGTGACTGCGTGTGAATTATTTTGTCAACTGAAGTTGAATGATTTGGCCAACTATATAGATAATCATTAATGGCATTTGTT-----------TT----T-----------------------------------------------------------ATG-GTCAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCGACAACAGGTGAATATT----CC------------GCATCGGCTATCATGTCCAGGGCCTCGGTGTGTCGACAT |
| dp5 |
XL_group1a:5757281-5757551 + |
|
GTCTCCAC----------------A--------------------------CTCCATCAACACACCCACACTCA-----------------------------------TGC------------CACATAAGC--------------------------------------CACACACCCA--------------------------------------------------------------------------------CACAAACATAT----------------------------AAACACGCATACATA---CATACATACATACATATATGGCT----A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTATATGGCAGCCCG----------ATCTGCCTTTTAGTTTTTAGTTCA---CAATTGTTTTTT-TGTGTATGT-ATAAAATATTTGTA--A----------------TTAATTATTTTGT----------------AAACGAACCTC----------------------------------------------------------------------------C-----------------------------------T---------------------------------------------------------------------------------------CTCCACACACAAA--CACA--------------------------------------------------------------------------------------------------A------GCGCACATACACATACCAACA------CGCACACACA------------------------CACACACACA----------------------------------------CACACA-------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_0:4416990-4417285 + |
|
CA-CACAC----------------------------------------------------------------------------------------ACACACACACACGCACACAAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATACATTT---------------------AAGCCAATT--------------AGCATTTAT-----------------------------------------------------------------------------------------------------------------------------------------------GTGCGACGCTGGGTG---------------------------------------------------GCAA----------------------------------------------------------------------------------------GGGGCCGCAGGAGCAC-------------AAAATGCGTTACATTT----------T------CAT----------------------------------------CAAACGCA------------CGACACTTTGT------------------TTTAATTTGAGCAACCG----CACG----CACAC-----------------------------------------------------------A-----------------------------------TA-------------------------------------------CAC-------A----CATGGTTACAGTCACCCCCTCACAGAC--A------------------CCTACACACG------CATACATA--------------------------CATACACAGTGACAGGCTTTGGTTTGGTCTCGTCACGGTT--TGGAGG------C----------------CACATTCTCTCGGGCGCTCACGCATTTAC--------------------------------AGATTTT---------------------CT---------------------------------A |
| droAna3 |
scaffold_13335:2179882-2180129 + |
|
ACTCACAA-----------------------------------------------------------GGATTCAACGACAAGGACA-----------------------------------------------CCC------------------------------------ACACACACA--------CACACACA----------------------------------------------------------CACACACACACA-----TACTTGACGACGCGTTTACATATATCACGGAAACA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACAAA----------------------------------------------------------------------------------------GCCATTGATACAA-------------AATATATG-------------------------------------------------------------------------------------------TTATATAT------------------ATAAAATA-------------T--ATAT---------------------------------------------------------------------------------------------------------------------------------------------------------ATCT------------------------------------------CACACACACACACAGACACACACACAGACACACACACACAGACACAAAGA---------ACATACAAACTAACCGAT----------------------T--TATATA----------------------------------------------------CT---------------ATATTATTAATATGTATATATG----CC--------------------------------------------------- |
| droBip1 |
scf7180000396575:95888-96173 - |
|
ATACATTA----------------------------------------------------------------------------------------ACTTATACGTACGTATATGTAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATATGTGG---------------------GTTC-----------------------------------------TAT---------------------------------------------------------------------------------------------------------------------------ATATGTATACCTAGGGAAA----------TATAGATCGTCACTCTTATACATATACAC-------------------------------------------------------------------------------------------------------------------------------------AAAATATGCCAAATGG----------T-----ATAT----------------------------------CGGGGATTATCACACAACATTTAGTTCAAAAAATCAA------------------TATATATGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTACTCACTCCCTCACA--C------------AC--AC------ACACACACACACACTCTAGCA------GACACACACA------------------------CATACAAACAGA------------------------------CATATT--AAGGAT------------ATA-----------------------------TATTGCCTTT--------------CGATCCGCAAATGTT----CG------------GATTTGAATATTTTGT----------------------- |
| droKik1 |
scf7180000302610:1714411-1714697 - |
|
CA-CACAG----------------------------------------------------------------------------------------ACACTCCCACACACACATACAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACACTTAC-------------------GGAC----AGCGGCAGACAAAGCCGAGCAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAAAC-AACATTGAAAAGTCTGTCTCTCTGTCTGCCACGCG---TGGTTGTGTGTG-TGTGTGTGT-GATAGACAGGCGTAAAG----------------TTAATTAAACA--------------------------------------------------------------------------------------------------------------------------------ATAAATA-------------C--ACACACATAC-----------------------------------------------------------ACA-TGCAGAC----A----------------------------------------------CCAGC--------------------------CGACACCGACACA--G------------GC--AC------CCGCACACACCCACATACACA------CACACACACA------------------------CACACACGCAGA------------------------------CGCGTT----------------------------------------------------A---------------AAAATGGCGAAAAACTACGGACT----AC------------ACT-----------------------------------T |
| droFic1 |
scf7180000453829:1700821-1701103 - |
|
TCGC--------------------------------------------------CGTCTACATGCATACATCTA-----------------------------------TGG------------TGCTGTGTG--------------------------------------TATTCATTTA--------------------------------------------------------------------------------CACACACACAT----------------------------GTATACACGTACATG---TATACATAC----------------------------------------------------------------ATATATGTAT---TAA-------------------ATA-----TCTCCGCTTTTTTTACGGCGCAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AG-----TGGCGCGTGAATTGTTTTTGCAC----AGTTGGGCG------------------------AA----------T--ATAACGCATT----CGCAT----------------TCACACGCACGCCGTCCCACAAACAAAGGAC----------------------------------AAAA--CGCGCATACG-------------------------------------------CCC-------A----GTAGGACAAACAAAACTCTCCG----------------------TGTGCCCACACACA------CACACACACA------------------------CAAACACATA----------------------------------------GACACA-------------------------------------------------------------------------------------------------------------------------------------------- |
| droEle1 |
scf7180000491087:568184-568426 + |
|
ACACACAC----------------------------------------------------------------------------------------TCACACACACACATATATAAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGACACAGAT---------------------ACACAGGTA--------------CCCACACTGATGTTTTCAAATGTTTCAGCATTTTTACAC---ACA---------------------------------------------TCTCA---------------------------CACGTCGAAATCCCCGC------------GGAATAGCAAGTCAAACGGAATACCAGAAAGGGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCACCCCA---------------------------------------------------------------------------------------------AATCACACAGACACGCACACAC------------------AACA--CCCACACAGG-------------------------------------------CCC-------A----CTGGGA----------------------------------------TC------CACA------CACACACAC--------------------------ACACACACACA----------------------------CACATCCT----------------------------------------------------CG---------------CTATT-------------------------------------------------------------------------- |
| droRho1 |
scf7180000777040:4962-5201 + |
|
CA-CACAC----------------------------------------------------------------------------------------ACTCACTAACAGACAC-----------------------------------------------------------CACACACACA--------------------------------------------------------------------------------------------------------------------------------CACACA---------------------------------------CACACTCACATACAC------------------------ACATGCAC------------------------------------------------------CG------------------------------------------------------------------GGGGAA----------TACATGTGA---------------ATGAGGGGCCCAGATGAGCGTTCATAAT--------------------------GTGT-------GC-TTAATTATCGCAGAGATATATGTA--TAT-------------------------------------------------------------------------------------------------------------------------------------------ACTACCCCC------------------AA----------C--CCAACACACA----CACAC-----------------------------------------------------------ACA-CACACACACACACACACA--CACA--------------------------------------------------------------------------------------------------C------ACACACACACACACACACACA------CACACACACA------------------------CACACACACA----------------------------------------CACACA-------------------------------------------------------------------------------------------------------------------------------------------- |
| droBia1 |
scf7180000301141:1075-1299 - |
|
GCCAGCGCCA-ACAGCATCGGCCGCCGGCTCCGTTCTCACATCAGCAGTAGCCACACACAC-----------------------------------ACACGCACACACACACGCACAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACACGCAC---------------------A--CACACACACACACACGCGCACACACAC--------------ACACACACACACACACACACA-C------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACACACA----C-----------------------------------------------------------ACA-CACACACACACACACACA--CACA--------------------------------------------------------------------------------------------------C------ACACACACACACACACACACA------CACACACACA------------------------CACACACACA----------------------------------------CACACA-------------------------------------------------------------------------------------------------------------------------------------------- |
| droTak1 |
scf7180000414113:33879-34129 - |
|
CA-TATGT----------------------------------------------------------------------------------------ATATATGTACATATGTATATAT----------------------------------------------------------------------------------------------------------------------------------------------------A-----C-----------------------------------CGTTGAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA----------TGCATACTA---------------CTTATAGTTTTTTAAAGTTATTAATTAT--------------------------TTTT-------ACCGTTATTGTTGTTGTTGTTGATGT---TAT-------------------------------------------------------------------------------------------------------------------------------------------ATTATTGTT------------------GT----------TGT-----------------------------------------------------------------TGCTGACATGCACATA-CACACAC------ACACA--CACA--------------------------------------------------------------------------------------------------T------TTGTACA--------------------CACACGCACA--------CCAAGACACAATCACACATACGATCACA------------------------------CTT--TG-----CA-------------------------------------------------ACTCTTTTTTGGCAATTGCTAACATTGGAATATC----TC------------TCTCT---------------------------------G |
| droEug1 |
scf7180000409711:3150997-3151270 - |
|
CCGCCGCC----------------------------------------------------------------------------------------GCCCACTGTCACGCACTCCTGCACCCGCCCCTTAC--CCC-AAAAATAACCCCATCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATGGGTATATCTATGTTATAGGTGTATGTAG----------------------------GTATGTATGTTTGTATGTACACCTATGGGGA----------CAA----------------------------------------------------------------------------------------------------------------------------------ACAGCCGAG-------AT---------------------------AAGCAAACACT-----------------------------------------------------------------------------------------------------CCTCG-------------C--ACACAC--------------------------------------------AACAAACACAC-----------------------A----------------------------------------------C--CTGGGCCACATCGATGCTTATGAGTTGTGCA------------------------------CACATAAAC----ACATA------CACA------CACACACACA------------------------CACACACACATA------------------------------C----TTATCCATA---------------------------------------------A--------------GTAATG-GCAAAAGGTG--------------------------------------------------------------- |
| dm3 |
chrX:6784128-6784486 - |
|
ACACAAAC----------------------------------------------------------------------------------------ACACACAAACACACACACACAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACGCACTT---------------------ACCTGGGCA--------------TGTGT-----------------GACTCAAA---T---------TATATATAATTTCATTGGAGC--ATATTTTTCCTGTTGTGATTTCTGCCTCG-------------ACTAGGATGTTTCT--TGCCCTTTTCCAGCCAGGATGTGTGAAGGCGCAGCGAATGGGTTTTATGGCCAGGATGTCTGTCACCTTCACACATACACAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCCACACTCA----C----------------------------------------------------GAGGGACTCAC-----------------------------------------------------------------------TCGC--------------------------TGACTCCGACAGA----------------CACAC------------ACACA------CACA------CACACACACACACGCA------------------CACGTACACA----------------------------------------TGCACATGGCTAGTCAGA---GGCATACT---------------------T---------------CAATTAGC--------AGATAAT----TT------------ACTT------------TGGACTCATCAGAGT-TCATCAG |
| droSim2 |
3r:9211992-9212204 - |
|
TTCGATG----------------------------------------------------GGCTGCACATATACACCG---------------------------CCATGTACACATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACC-CGGTAAG---CATACACAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACACAGCAACAAACAGCGACAAAGCATTA-A----------GCTTTCATGTGAGTGTGTTTGTAGCCGTGAGCGTGTCTG-TGTGTGTGC---GAA-------------------------------------------------------------------------------------------------------------------ACTTTTATTGTCAG---------------------------------------------------------------------------------------------------------------------CACAAACAGAC--AC-------------------------------------------------------------------------------------------------------A--------------------A------AC----TCACACACA------------CACACACACACACAAA------------------CACAAACACA----------------------------------------TACACA-------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec2 |
scaffold_12497:79-315 + |
|
ATATACAT----------------------------------------------------------------------------------------ATACATATACATATACATATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATACATAT---------------------ACATATACA--------------TATACATATACATATACATATACATATACATATACATAT---ACA----------------------------------------------------------------------------------------------------------------------------------------------------------TAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACATATA----CATATACATATACATATACATA--------------------------------------------------------------------------TACATATACATATACATATACATATAC--AT-----ATACATATACAT-------ATACA----TATACATATACATATACATATAC------------------------------------------------------------------------------------------------------------------------------------------A---TATACATATACATATACATATACATATAC--------------------------------ATATACA----TT------------TGTCG---------------------------------A |
| droYak3 |
X:19121601-19121823 + |
|
GACACCGC----------------------------------------------------------------------------------------ACACAGGACTCCAAAGGTATAC--------------------------------------------------------ATATATGTATGTACGTATGTACATATGTATGTATGCATGCCGCTCTGCGAAAATTGCACGAAAATTGGAAATTTATTGCCAAGCCCACACAAACACAT----------------------------AAAAGCACAGGCAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GACACA----C-----------------------------------------------------------AAG-CACACTCACACACACAAG--CACA--------------------------------------------------------------------------------------------------C------TCACACACACTCACACACACG------CACAAACACA------------------------CACACACACA----------------------------------------CACACA-------------------------------------------------------------------------------------------------------------------------------------------- |
| droEre2 |
scaffold_4845:7337556-7337805 + |
|
ACATACAT----------------------------------------------------------------------------------------ACATACATACATACATACATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACATACAT---------------------ACATACATA--------------CATACATACATACATACATACATACATACATACATACAT---ACA----------------------------------------------------------------------------------------------------------------------------------------------------------TAC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACATA----CATACATACATACATACATACA--------------------------------------------------------------------------TACATACATACATACATACATACATAC--AT-----ACATACATACAT-------ATATA----TACATACATACATACATACATAC------------------------------------------------------------------------------------------------------------------------------------------A---TACATACATACATACATACATACATACAT--------------------------------ACATACATACACCGGTACATACACCGT------------------------------------T |