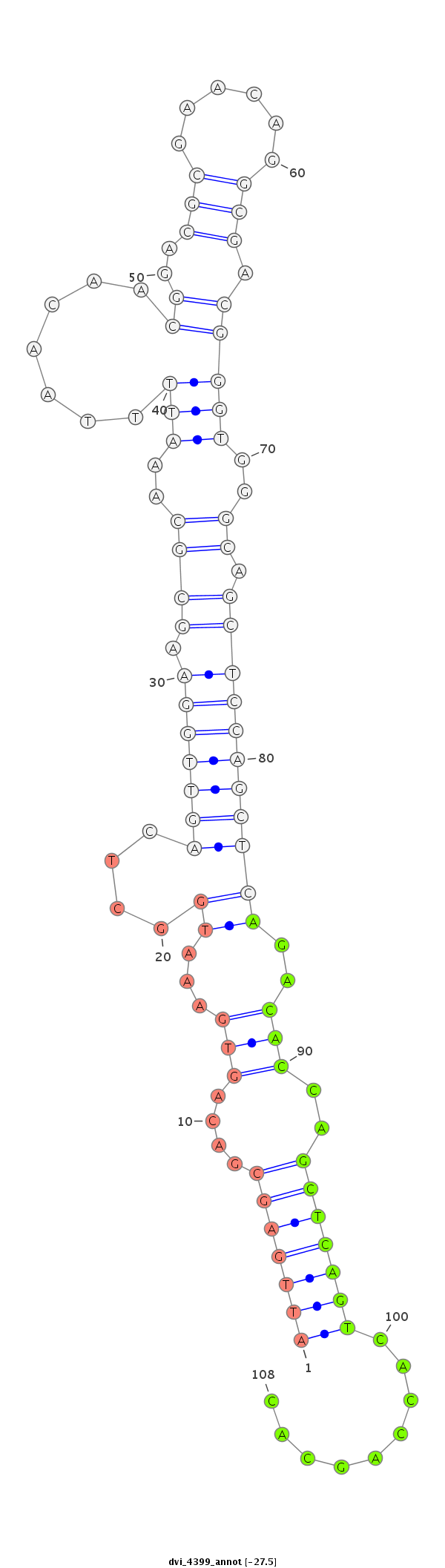
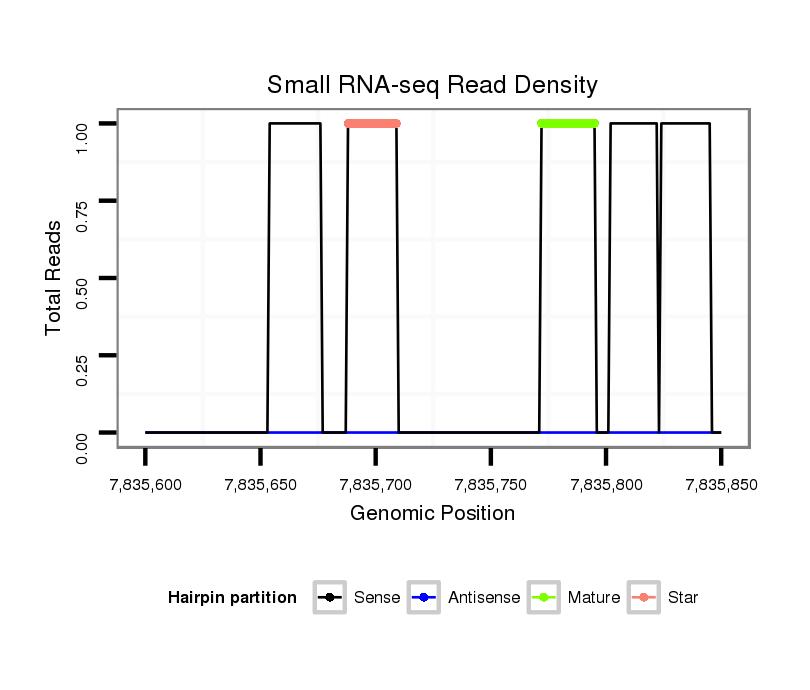
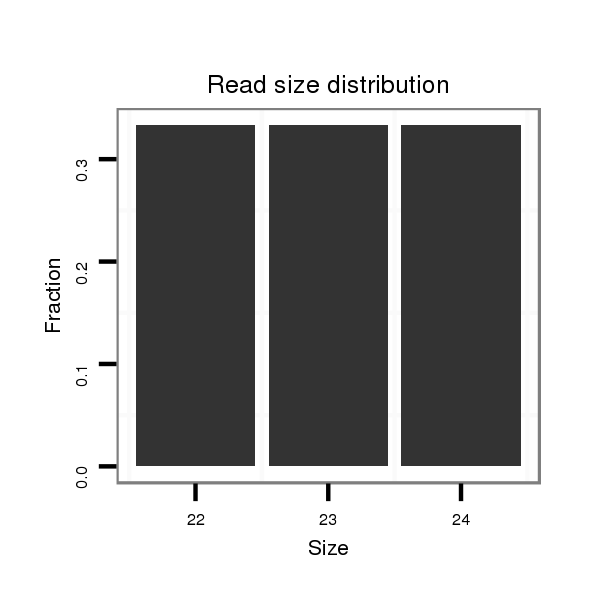
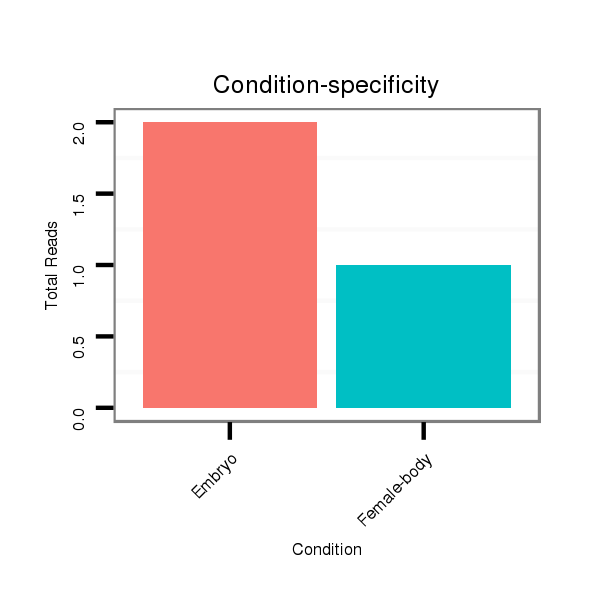
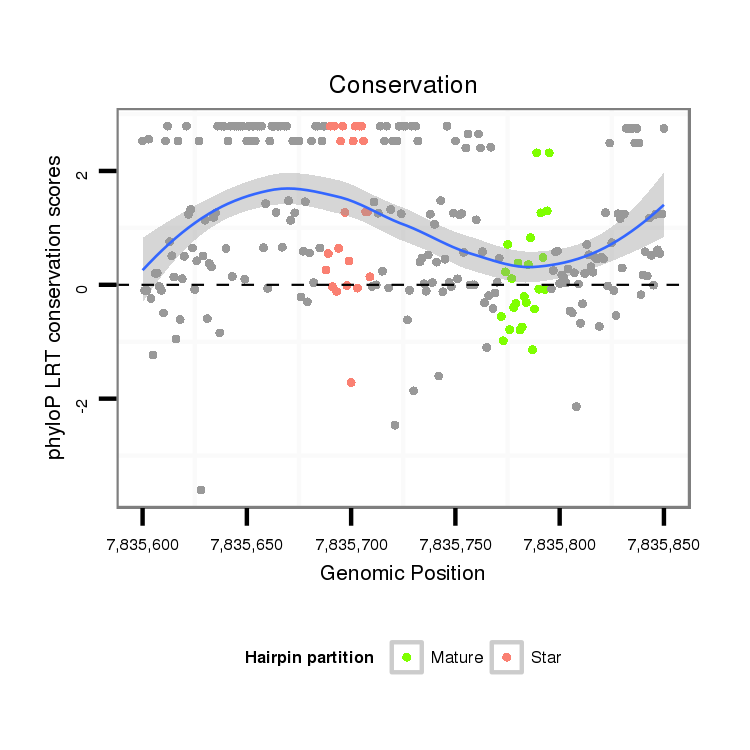
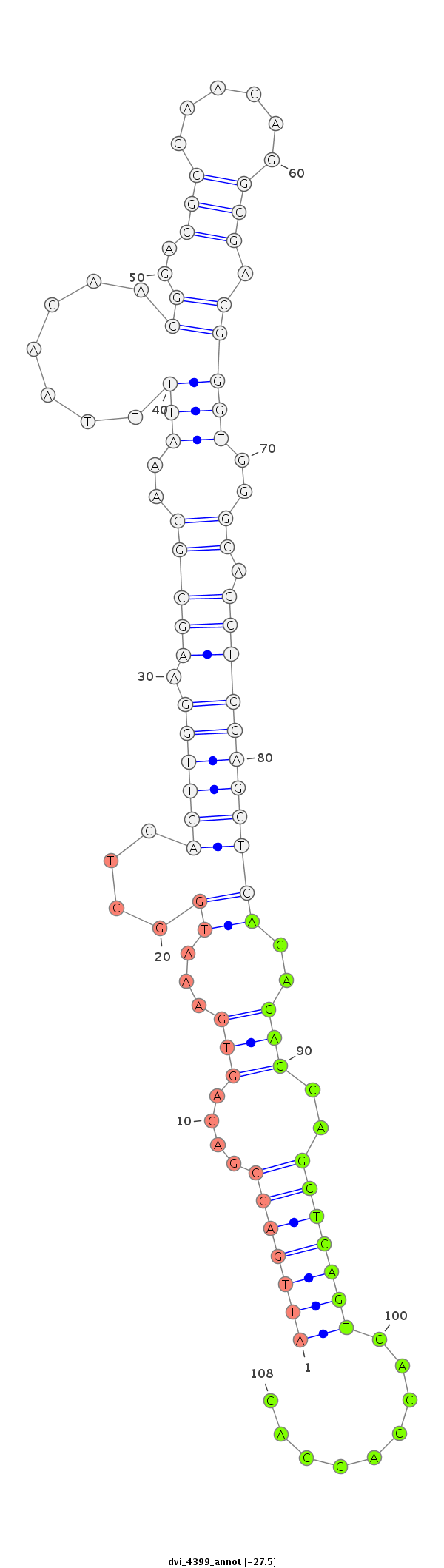
ID:dvi_4399 |
Coordinate:scaffold_12855:7835650-7835800 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.5 | -27.4 | -27.4 |
 |
 |
 |
exon [dvir_GLEANR_10902:2]; CDS [Dvir\GJ11017-cds]; intron [Dvir\GJ11017-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CCCTGGCCGTCGATACCGAGCAGCGCTCCAAGCTGCTCAACGACAAATACGTGAGTAACAGCATCTTCAATGGCGGGCAGCGGAACGAATTGAGCGACAGTGAAATGGCTCAGTTGGAAGCGCAAATTTTAACAACGGACGCGAACAGGCGACGGGTGGGCAGCTCCAGCTCAGACACCAGCTCAGTCACCAGCACCAGCACAACAGTTATACCCTCACCAGCGGAACCCGCTAATGAGCCTAGCACATCA ****************************************************************************************(((((((....(((...((....(((((((.((((..(((.......((..(((......))).)))))..)).)))))))))))..)))..))))))).........******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060666 160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
M061 embryo |
SRR060672 9x160_females_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
V047 embryo |
SRR060671 9x160_males_carcasses_total |
V053 head |
SRR060685 9xArg_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
M027 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060689 160x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060678 9x140_testes_total |
SRR060681 Argx9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060670 9_testes_total |
SRR060674 9x140_ovaries_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................GGGGGCAACTCCATCTCAGA............................................................................ | 20 | 3 | 5 | 53.40 | 267 | 119 | 1 | 24 | 14 | 15 | 13 | 10 | 12 | 12 | 8 | 4 | 5 | 2 | 0 | 7 | 0 | 5 | 3 | 3 | 2 | 3 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ACATGGCGCAGTTGGTAGCG................................................................................................................................. | 20 | 3 | 14 | 9.36 | 131 | 1 | 82 | 23 | 0 | 0 | 2 | 2 | 3 | 1 | 3 | 0 | 6 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GGGCAACTCCATCTCAGA............................................................................ | 18 | 2 | 4 | 6.25 | 25 | 11 | 0 | 2 | 2 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GGGCAACTCCATCTCAGAG........................................................................... | 19 | 3 | 18 | 3.28 | 59 | 3 | 9 | 8 | 2 | 4 | 1 | 6 | 8 | 0 | 1 | 0 | 3 | 4 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................AGTGGGCAACTCCATCTCAGA............................................................................ | 21 | 3 | 2 | 3.00 | 6 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ACATGGCGCAGTTGGTAGCGC................................................................................................................................ | 21 | 3 | 3 | 1.67 | 5 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................AGACACCAGCTCAGTCACCAGCAC....................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GTGGGCAACTCCATCTCAGACC.......................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AACAGTTATACCCTCACCAGC............................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GGGCAACTCCAGCTCAGA............................................................................ | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATTGAGCGACAGTGAAATGGCT............................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GAACCCGCTAATGAGCCTAGCA..... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GTAACAGCATCTTCAATGGCGGG.............................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................CAATGGCGGGGAGCGGACC..................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................ATAGGCTACGGGTGGGCTGCT...................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GTGGGCAACTCCATCTCAGAG........................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GGGGGGCAACTCCATCTCAGA............................................................................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATAGCTCAGTTGGTAGAGCAAA............................................................................................................................. | 23 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GGCGGAATCCGCTAAGGAGC........... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GGGGGGCAACTCCATCTCAG............................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CGGAATCCGCTAAGGAGC........... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GAAATAGCTCAGTTGGAAG................................................................................................................................... | 19 | 1 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GAAATGGCTCAGTTGGGAG................................................................................................................................... | 19 | 1 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AGCAGGGCTCCAAGGTGCACA.................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................ACAGTGAGATTGCTCAGT......................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................TGATATGGCTCAGTTGGTA.................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GCGGGCAACTCCATCTCAGA............................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AGTGGCTCAGTAGGAAGAGC................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATGGCGCAGTTGGTAGCGC................................................................................................................................ | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GGGCAACTCCATCTCAGAGA.......................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................CACATGGCGCAGTTGGAAGC.................................................................................................................................. | 20 | 3 | 14 | 0.14 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................GCGCTTCAAGCAGATCAACGA................................................................................................................................................................................................................ | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GAAATAGCTCAGTTGGTAGCG................................................................................................................................. | 21 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................GAAATGGCTCAGTTGGGA.................................................................................................................................... | 18 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................GAAATAGCTCAGTTGGAAGTG................................................................................................................................. | 21 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GAAATAGCTCAGTAGGAAG................................................................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ACATGGCGCAGTTGGGAGC.................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GGCAACTCCATCTCAGAGA.......................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACGACAAATATGTGTGTAAGA............................................................................................................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................AGGGGGCAACTCCAGCTCAG............................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................GAAATAGCTCAGTTGGAGGG.................................................................................................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GAAATGGCTCATTTGGGA.................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................GACATAGCTCAGTTGGTAGC.................................................................................................................................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CGAAACCGAGCGGCGCTAC.............................................................................................................................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ACGGCTCAGTTGGTAGAGCA............................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GAATTGGCTCAGTAGGGAG................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ACATGGCACAGTTGGTAGCG................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GCTCAGTTGGTAGAGCAACT............................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GACAGTGAAACGCCTTAGT......................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GGGGCATCTCCATCTCAGA............................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GATATGGCTCAGTTGGTAGAG................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GGGACCGGCAGCTATGGCTCGTCGCGAGGTTCGACGAGTTGCTGTTTATGCACTCATTGTCGTAGAAGTTACCGCCCGTCGCCTTGCTTAACTCGCTGTCACTTTACCGAGTCAACCTTCGCGTTTAAAATTGTTGCCTGCGCTTGTCCGCTGCCCACCCGTCGAGGTCGAGTCTGTGGTCGAGTCAGTGGTCGTGGTCGTGTTGTCAATATGGGAGTGGTCGCCTTGGGCGATTACTCGGATCGTGTAGT
*******************************************************(((((((....(((...((....(((((((.((((..(((.......((..(((......))).)))))..)).)))))))))))..)))..))))))).........**************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M047 female body |
V116 male body |
SRR060686 Argx9_0-2h_embryos_total |
M027 male body |
SRR060665 9_females_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060680 9xArg_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................GTCGTAGAAGTTACCGGGC.............................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTGCTGCACTCATTGTGGTAG.......................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....CGGCAGCTATGGGTCGTCG................................................................................................................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TGCTGCACTCATTGTGGTAG.......................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................TCGAAGAGGTGCTGTTTA........................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GTCGTGTCGTCAACATGGGG................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................GCTGCACTCATTGTGGTAG.......................................................................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................GCGGTTGTCAGCTGCCCA.............................................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................CGAGACAGTGGGCGTGATC.................................................... | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................AGTCAGTGGGCGGGGTC.................................................... | 17 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GTCGTAGTGGTTACTGCCC.............................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:7835600-7835850 + | dvi_4399 | CCCTGGCCGT------CGATACCGAGCAGCGCTCCAAGCTGCTCAACGACAAATACGTGAGTAACAGCATCTTCAATGGCGGGCAGCGGAACGAATTGAGCGACAGTGAAATGGCTCAGTTGGAAGCGCAAATTTTAACAACGGACGC------------------------GAACAGGCGACGGGTGGGCAGCTCCAGCTCA-----------------------------------G---------------A---------------------------------------------------CACC----------------AGCTCAGTCACCAGCACCAGCACAACAGTTATA------CC-----CTCACCAGC------------GGAACCCGCTAATGAGCC---------------TAGCACATCA |
| droMoj3 | scaffold_6540:23056093-23056373 + | dmo_2032 | CTCTGACCTTCGAGGCCGATCGCGAGGAGCGGGCCAAGGCGCTCAACGAGAAATATGTGAGTAACACCATCTTCAATGGCGGGCGACGCAACGAGTTGAGTGACAGCGATATGGCTCATCTGGAAGCCCAAATATTGACAAACGA---------------------------AAACAAGCGACGGGT---TAGCAGCAGCAGCA--GCAGCAG------C------GG---------------------------------------------------------------------CAGGGGCAGTGGCAGC---TCAGTAAGCACCACCAGCACCAGCAACAGCCCCACAGCTCTGG------C------CCCACAAGT------------GGTGCCCACTAATGAGCC---------------CGCAATGTCA |
| droGri2 | scaffold_15074:4400745-4400959 + | CGCTATCCAGCGACATGGAGCCGGAGCAGCGCTCGAAGGCGCTCAATGACAAATATGTGAGTAACAGCATCTTCAATGGCGGCAAACGCAACGAACTCACCGACACTGAAATGGCCAACCTAGAGGCACAAATATTGACAACGGTGCC------------------------CAGTAGACG---------------------------------------------------------------------------------------------------------------------------------------------------------ACCCACAACAACCAGCAGCCCCCCG------C------TGA-----C------------CGAGGCCACTAATGAGCG---------------CACAACACCA | |
| droWil2 | scf2_1100000004943:12858435-12858683 + | CGA--ACAAT------TGATGATGATGAGCGGTCGAAGGTGTTGAATGAGAAATATGTGAGTAACAGCATCTTTAATCGTGGCCAACGCAATGAGTTAAGCGACCAAGAAATGGCCCGCCTGGAAGCGCAAATTTTAACAGGAATCGGTTCTTCTTTGGCATCGGGTTCAAATAGCAAGCGAAAGGT---CAGCAAG---------------------------------------------------------------------------------------------------------------------------------------------------GTCAACAGCGGTAGT------C------GAT-----C------------TGAATCCACTAATGAGCCGACCACAACCAGCAGCACAACGGCA | |
| dp5 | 2:21717393-21717727 + | CTGTCTCCTC------GGATCCGGCGGAGCGGGCCCGGCTGCTGAACGAGAAATACGTGAGTAACAGCATCTTCAATGGCGGGCAGAGGAACGAGCTGAGTGACTCCGAAATGGCCAATCTGGAGGCCCAAATACTCACGGGCGTGGG---------------------GCCAACGAAGCGGAAAGT---CAGCCTGAATAAGTCCGCTGCTCCACCAGCCACCACCACTCCAGCTTCT---------------GCCTCTCCTCTGGCTGGTGATCCTGCTCCTGC------------------------------TGCTCCTGCCCTGTTGGCCACCATAACAGTCTCCGATTGGAAGCCAACGC------CAC-----CTGGGCCCGGGCCTGGGCCCACTAATGAGGC---------------CACAACGGCA | |
| droPer2 | scaffold_3:4502125-4502459 + | CTGTCTCCTC------GGATCCGGCGGAGCGGGCCCGGCTGCTGAACGAGAAATACGTGAGTAACAGCATCTTCAATGGCGGGCAGAGGAACGAGCTGAGTGACTCCGAAATGGCCAATCTGGAGGCTCAAATACTCACGGGCGTGGG---------------------GCCAACGAAGCGGAAAGT---CAGCCTGAATAAGTCCGCTGCTCCTCCAGCTACCACCACTCCAGCTTCT---------------GCCTCTCCTCTGGCTGGTGATCCTGCTCCTGC------------------------------TGCTCCTGCCCTGTTGGCCACCATAACAGTCTCCGATTGGAAGCCAACGC------CAC-----CTGGGCCCGGGCCTGGGCCCACTAATGAGGC---------------CACAACGGCA | |
| droAna3 | scaffold_13340:23256652-23256962 - | CACTCACCGGAGGCGAAGATCCGGGCGAGAAGGCGCGACTCCTCAACGAGAAATACGTGAGTAACCACATCTTCAATGGCGGGCAGAGGAACGAGCTAACCGACGCGGAAATGGCTAACCTGGAGTCACAGATTATTACGGGTCTGGG---------------------CCCAATGAAACGAAAGGT---CAGCCCAAACCAATCCGCTCCTG------C---------TCCGGCTCCG---------------ACTGCTC---CGCCTGCT---------------GAAGCTG---------------CAGCACCTTCACCGGCCATTCTCACCTCCATCACGGTCTCCAAGTGGA---------------CAC-----C------------AGAGACCGCTAATGAGCCTACCACCAC------CACCACGGCA | |
| droBip1 | scf7180000396395:258171-258493 - | CGCTCACCGGCGGCGAAGATCCCGCGGAGAGGGCGAGGCTCCTAAACGAGAAATACGTGAGTAACAGCATCTTCAATGGCGGGAAGAGGAACGAGCTAACCGACGCGGAGATGGCTAACCTGGAATCCCAGATTATCACGGGTCTGGG---------------------CCCAATGAAGCGAAAGGT---CAGCCCGAACCAGTCTGCTCCAG------C---------TCCTGCTCCT---------------GC---TG---CTCCTGCTGCTCCTACT---GCCGCTG---------------CTGAAGC---TC---CGGCCATTCTCACCTCCATCACGGTATCCAAGTGGACACCG---A------CAC-----C------------GGAGTCCGCTAATGAGCCTACCACCACCC---CCGCCGCGGCA | |
| droKik1 | scf7180000302707:169940-170247 - | CGGTTGGTGG------AGAACCGGAGGATAAGGCACGGCTGCTCAACGAGAAATACGTGAGTAACAGCATTTTCAATGGCGGTCAACGAAACGAGCTGACCGATGCGGAAATGGCTAATCTAGAATCCCAGATTCTGACTAACCTGGG---------------------CCCCATGAAACGGAAGGT---CAGCCTGACTAAATCCGCTCCTG------C------TGCTCCTTCT------------------------GCTGCTGCTGGGGCTTCTCCT---TCCGC------------TGATTCCGACGC---TCCT------ATATCAACGTCCATAACAGTCACCAAATGGACT------CCGCCAGCAC-----C------------TGAATCGACTAATGAGGC---------------CACCACGGTA | |
| droFic1 | scf7180000453850:442311-442606 - | CGGTGGCCGGGGATCCGGATCGGGCGGAGAAGGCTCGCCGGCTCAACGAGAAATACGTGAGTAACAGCATCTTCAACGGCGGCCAGAGGAACGAGCTGAGCGACGCCGAGATGGCCAATCTGGAGTCCCAGATTCTGACTAACCTGGC---------------------TCCCGCGAGGCGGAAGGT---CAGCCCCAGTCAGTCCGCTCCT---------------------GCTGCC---------------GCATCTC---CCCCTGCT------------------------------GACTCTGCGGC---TCCTGTTGCCATGACCGCTACCATAACGGTCACCAAGCTCAGT------C------CAC-----C------------TGAAGTCACTAATGAGAC---------------CGCCTCGGCA | |
| droEle1 | scf7180000491104:2898467-2898765 - | CGTTGACCGG------CGATCCGGCGGAGAAGGCACGTCTGCTCAACGAGAAATACGTGAGTAATAGCATCTTCAATGGCGGTCAAAGGAACGAGCTGAGCGACACCGAGATGGCCCATCTTGAGTCCCAGATTCTGACTAACCTGGG---------------------CCCCATGAAAAGGAAGGT---CAGTGTGACCAAGTCCGCTCCTG------C------GGCTCCTGCTGCT---------------GCCGCTT---CTCCTGTT------------------------------GATTCCGCAGC---TCCTGTTGCCATAGACGCTTCCATTACGATGACGAAGCTGAGT------C------CAC-----C------------TGAAGCCATTAATGAGAG---------------CGCCCCGGCA | |
| droRho1 | scf7180000777712:2540-2838 - | CGGTGACGGG------AGATCCGGCGGAGAAAGCTCGGCTGCTTAACGAGAAATACGTGAGTAACAGCATCTTCAATGGCGGTCAAAGGAACGAGCTGAGCGACGCAGAGATGGCCAATCTTGAGTCCCAGATTCTTACTAACCTGGG---------------------CCCTATGAAACGAAAGGT---CAGCCTGACTAAGTCCGCTCCTG------C------GGCTCCTGCTGCG---------------GAGGCTT---CTCCTGCT------------------------------GAATCCGCAGC---TCCTGCTGCCATGTTTGCTTCCATAACAGTCACCAAGCTAAGT------C------CAC-----C------------TGAAGCCACTAATGAGAC---------------CGCCCCGGCA | |
| droBia1 | scf7180000302411:817779-818092 - | CGGTGACCGG------AGATCCAGGGGAGAAGGCTCGGCTGCTCAACGAGAAATACGTGAGTAATACCATCTTCAATGGCGGTCAAAGGAACGAGCTGAGCGACGCAGAGATGGCCAATCTGGAGTCTCAGATACTGACTAACCTGGG---------------------ACCTATGAAGCGGAAGGT---CAGCGCGACCAAAACCGCTCCGG------C------GGCTCCTGCTGCT---------------GCGCCCT---CTCCTGCTGCGCCCGCTCCCACT---------------GATTCCGACGC---TGCGGTCGCCATAACCGCCTCTATAACAGTCACCAAGCTTAGA------A------CAC-----C------------TGAAGCCACTAATGAGGC---------------CGCCCCGGCA | |
| droTak1 | scf7180000415789:846247-846542 - | CGGTGTCCGG------AGATCCGGCGGAGAAGGCGCGGCTGCTCAACGAGAAATACGTGAGTAATAGCATCTTCAATGGCGGTCAAAGGAACGAGCTGAGCGACGCCGAGATGGCCAATCTGGAGTCTCAGATACTGACTAACCTGGG---------------------ACCTATGAAGCGGAAGGT---CAGCCTGACCAAATCCGCTCCAG------C------GGCTCCTGCTGCC---------------GCGTCCC---CTCCTGCTGCGTCT---------------------------------GC---TCCTGTGGCCATAACCGCTTCAATCACAGTCACCGAGCTTGGT------C------CAC-----C------------TGAAGCCACTAATGAGGC---------------CGCCACGGCA | |
| droEug1 | scf7180000409759:1037476-1037768 - | CGGTGAACGG------AGATCCAGGAGAGAAAGCACGTCTGCTGAACGAAAAATACGTGAGTAATAACATCTTCAATGGCGGTCAAAGGAACGAGCTGAGCGACGCAGAGATGGCCAATCTGGAGTCCCAGATCATTACTAACCTTGG---------------------GCCCATGAAGCGGAAGGT---CAGCCTGACCAAATCCGTTCCAA------C---------------TGTT---------------GCGTCCC---CTCCTGCT---------------GCAGCTGCTCCTAGTGATACCTTGGC---TCCTGTTG------------CCATAACAGTCGTCCAGTTAGGT------C------CAC-----C------------TGAAGCCACTAATGAGAC---------------CGCCACGGCA | |
| dm3 | chr3R:1069281-1069609 - | CGGTGGCCGG------AGATCCGGCAGAGAAGGCGCGACTGCTTAACGAGAAATATGTGAGTAATAGCATCTTTAATGGCGGTCAGAGGAAAGAACTGAGCGACGCAGAGATGGCCAATCTGGAGTCACAGATACTCACTAACCTGGG---------------------ACCTATGAGGCGGAAGGT---CAGTCTGACCAAAACCGCTTCAG------C------GGCTCCTGTTCGT---------------GCACCTT---CTCCTGCTGCTCCTTCTCCTGCCGAGGCTGCCCCTCTGGATTCGGCAGC---TCCTGCTGTATTAACCGCATCTATAGCAGTCACCAAGCTTAGT------C------CAC-----C------------TGACGCCACTAATGAGAC---------------GGCCACGGCA | |
| droSim2 | 3r:975974-976302 - | dsi_28105 | CGGTGGCCGG------AGATCCGGCAGAGAAGGCCCGACTGCTTAACGAGAAATACGTGAGTAATAGCATCTTTAATGGCGGTCAGAGGAAAGAACTGAGCGACGCAGAGATGGCCAATCTGGAGTCACAGATACTCACTAACCTGGG---------------------ACCTATGAGGCGGAAGGT---CAGTCTGACCAAATCCGCTCCAG------C------GACTCCTGTTCCT---------------GCACCTT---CTCCTGCTGCTCCTTCTCCTGCCGATGCTGCCCCTCTGGATTCCGCAGC---TCCTGTTGAATTAACCGCATCCATAACAGTCACCAAGCTTAGT------C------CAC-----C------------TGACGCCACTAATGAGAC---------------GGCCACGGCA |
| droSec2 | scaffold_6:1193130-1193458 - | CGGTGGCCGG------AGATCCGGCAGAGAAGGCCCGTCTGCTTAACGAGAAATACGTGAGTAATAGCATCTTTAATGGCGGTCAGAGGAAAGAACTGAGCGACGCAGAGATGGCCAATCTGGAGTCACAGATACTCACTAACCTGGG---------------------ACCTATGAGGCGGAAGGT---CAGTCTGACCAAATCCGCTCCAG------C------GACTCCTGTTCCT---------------GCACCTT---CTCCTGCTGCTCCTTCTCGTGCCGATGCTGCCCCTCTGGATTCCGCAGC---TCCTGTTGTATTAACCGCATCCATAACAGTCACCAAGCTAAGT------C------CAC-----C------------TGACGCCACTAATGAGAC---------------GGCCACGGCA | |
| droYak3 | 3R:1433973-1434274 - | CGGTGGCCGC------AGATCCAGCAGAGAAGGCACGACTGCTTAACGAGAAATACGTGAGTAATAGCATCTTTAATGGCGGTCAGAGGAAAGAACTAAACGACGCAGAGATGACCAATCTGGAGTCACAGATTCTCACTAACCTGGG---------------------ATCTATGAAGCGGAAGGT---CAGTCCTATCAAATCTGCTCCAA------C------GGCTCCTGTTCCT---------------GTGCCTT---TTCCTGCTGCTCCTTCTCCTGGTGAGGTTGACTCTCTGAATTCCGCAGG---TCCTGTTGCCCTAACCGCAACCATAACAGTCACAAAGCTTAGT------C------CAC-----C------------T------------------------------------------ | |
| droEre2 | scaffold_4770:1346478-1346821 - | CGGTGGTCGG------AGATCCGGCAGAGAAGGCACGACTGCTTAACGAGAAATACGTGAGTAATAGCATCTTTAATGGCGGTCAGAGGAAAGAACTGAGCGACGCAGAAATGGTCCATCTGGAGTCCCAGATATTCACTAACCTGGG---------------------ACCTATGAAGCGGAAAGT---CAGTCTGACCAAATCCGCTCCAA------C------GGCTCCTGTTCCTGAGCCTTCTCCTGCTGCCCCTT---CTCCTGCTGCGCCTTCTCCTGCTGAGGCTGCCCCTGTGAATTCCGCAGC---TCCTGTTGCCATAACCGCATCCATAACAGTCACCAAGCTTAGT------T------CAG-----C------------TGACGCCCCTAATGAGAT---------------GGCCACGGCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 10:13 PM