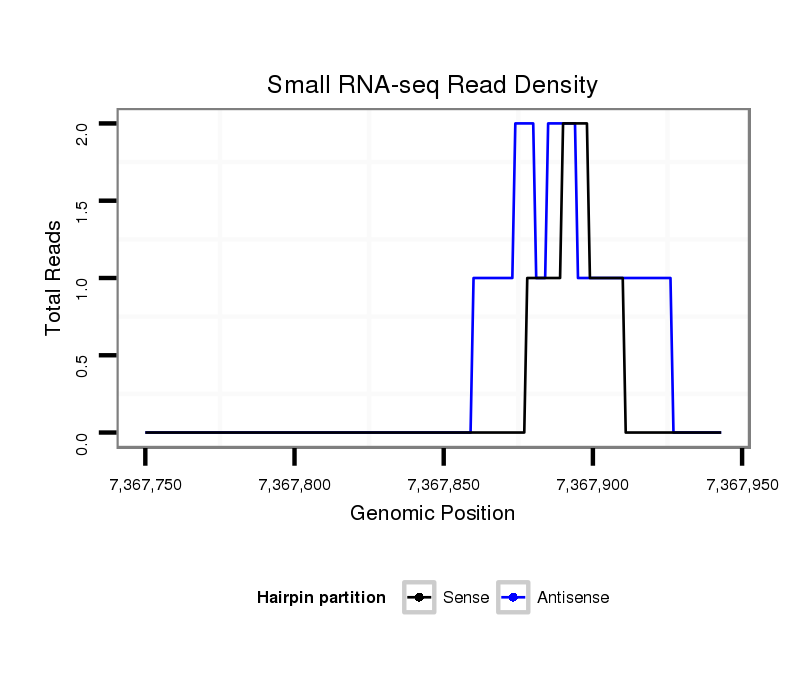
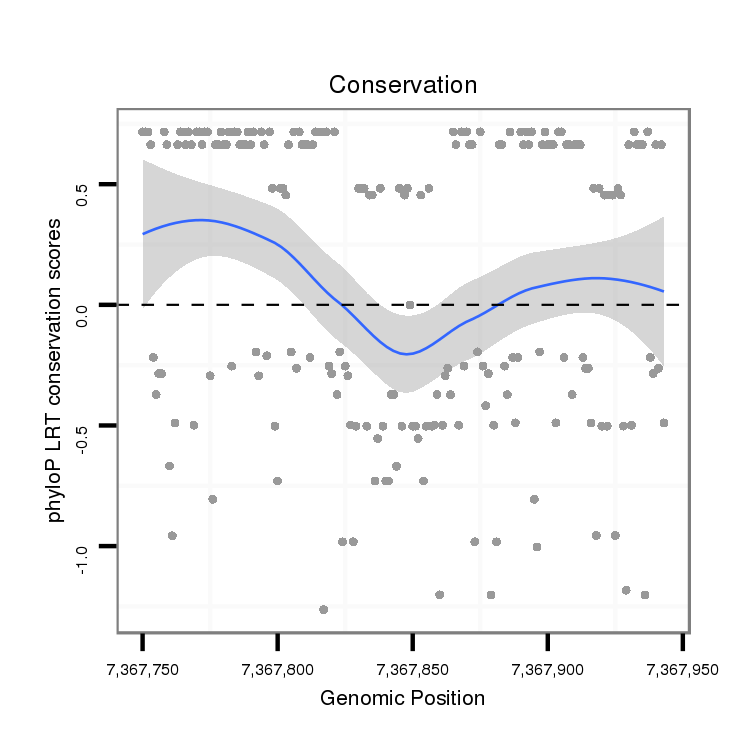
ID:dvi_4286 |
Coordinate:scaffold_12855:7367800-7367893 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_10876:1]; CDS [Dvir\GJ10990-cds]; CDS [Dvir\GJ10990-cds]; exon [dvir_GLEANR_10876:2]; intron [Dvir\GJ10990-in]
No Repeatable elements found
| --------------####################################----------------------------------------------------------------------------------------------################################################## AATGTCATTGCACCATGAGATACTTGCCCAGCAATTGCGACAGCTCTATGGTAGGGATTGGCGGTAACTATTGCCTCTCTTTACGGATTTGCCGGATGTTGCTGATAAACGACTGTCAATAGCCCAAATCACGCTGTTCCACAGACACGTCGCAATCGCCGGCATTGTCTAGACCCAGATGAACGCCATTCTGG **************************************************(((((((..((((((....))))))..)))))))(((...((..(((((((.(((((......))))).))))....)))..))...)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060681 Argx9_testes_total |
M061 embryo |
SRR060656 9x160_ovaries_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................TCACGCTGTTCCACAGACACG............................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................ACAGACACGTCGCAATCGCCG................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................AGCAATTGAGACGGCTCT................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................TAGACCATATCACGCTGTC........................................................ | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......TAGCACCATGAGATTCTT......................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TTACAGTAACGTGGTACTCTATGAACGGGTCGTTAACGCTGTCGAGATACCATCCCTAACCGCCATTGATAACGGAGAGAAATGCCTAAACGGCCTACAACGACTATTTGCTGACAGTTATCGGGTTTAGTGCGACAAGGTGTCTGTGCAGCGTTAGCGGCCGTAACAGATCTGGGTCTACTTGCGGTAAGACC
**************************************************(((((((..((((((....))))))..)))))))(((...((..(((((((.(((((......))))).))))....)))..))...)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060659 Argentina_testes_total |
SRR060683 160_testes_total |
V053 head |
SRR060655 9x160_testes_total |
SRR060662 9x160_0-2h_embryos_total |
M047 female body |
SRR060667 160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................GTTTAGTGCGACAAGGTGTCT................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................CGGCCGTAACAGATCTGGGT................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CAAGGTGTCTGTGCAGCGTTAG..................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CTGACAGTTATCGGGTTTAGT............................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TCTCTGAACGGGTAGTTAAC............................................................................................................................................................. | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TCGGTTTAAGTGCGACACGGT..................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................CGCAGTCGAGATACCGTC............................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................TATGAACGGGAGGTTAAC............................................................................................................................................................. | 18 | 2 | 19 | 0.11 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...................TCTGAACGGGACGTTAAC............................................................................................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| .............................TGGGTAACGCTGTCGAGAAA................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................GAGTTTAGTGCGACACGAT..................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:7367750-7367943 + | dvi_4286 | AATGTCATTGCACCATGAGATACTTGCCCAGCAATTGCGACAGCTCTATGGTAGGGATTGGCGGTAACTATTGCCTCTCTTTACGGATTTGCCGGATGTTGCTGATAAACGACTGTCAATA-----GCCCAAATCACGCTGTTCCACA-----------------GACACGTCGCAATCGCCGGCATTGTCTAGACCCA---GATGAACGCCATTCTGG |
| droMoj3 | scaffold_6540:25280627-25280822 - | AATGTTGCTGAGCCATGAGCTACTTGGCCAGCAGTTGCGACAGCTCTATATTAGGGAATGGCAGTAAGTGCTACTCCGTCTTATGGCATCTATATACGT-ATAGCCAGCTACCAATCCACA-----GCTCAGTCTCTGCCATTCTACA-----------------GAGCCGTCGCCATTGCTGGCAAAGTGTGGGCCGACGGGGGGCACGCTATCCAGG | |
| droGri2 | scaffold_15074:5492666-5492837 - | AATGATGCTGATGCATGAGATACTTTGCCAGCAATTGCGACAAATCCA------GAATTGGCGGTAAATACTATATATA-------------TAT-------------ATCAATATCAATATCTATGCATAATCGAAGCTATAGCACAAGCGCCGAAAGTGTTTGGAGTTGTCGCCATCGCTGGCTTTC---------------CGAACGCGAACCTGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:59 PM