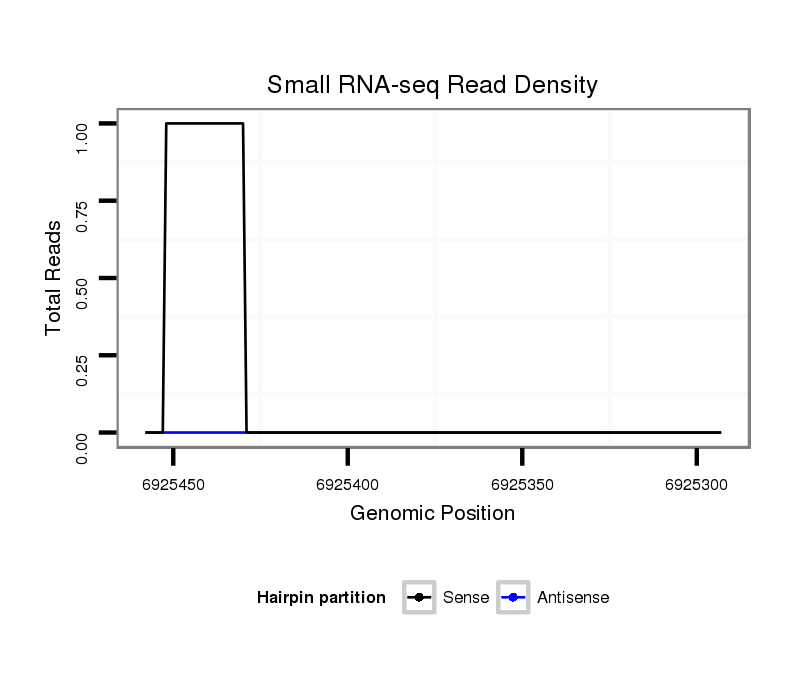
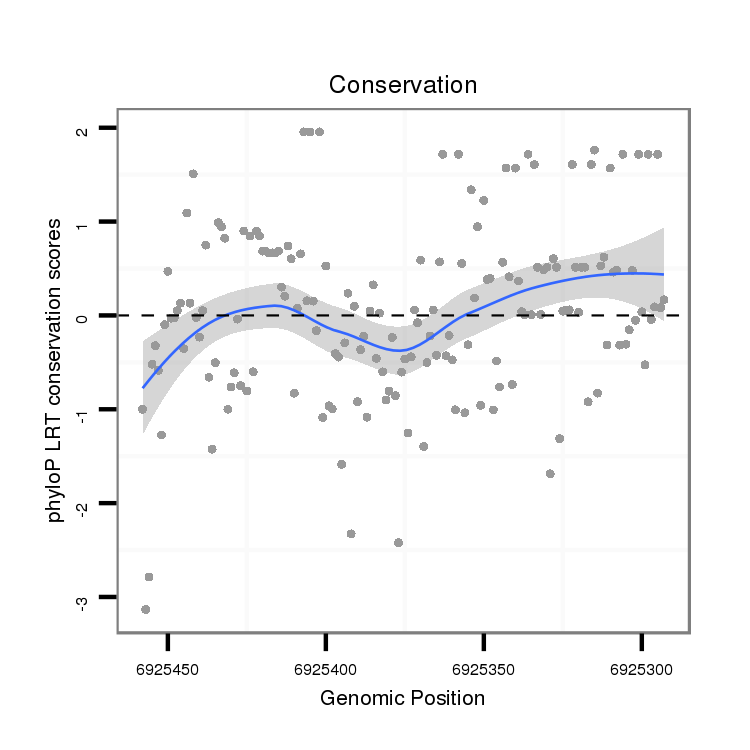
ID:dvi_4245 |
Coordinate:scaffold_12855:6925343-6925408 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_10141:5]; CDS [Dvir\GJ10201-cds]; CDS [Dvir\GJ10201-cds]; exon [dvir_GLEANR_10141:4]; intron [Dvir\GJ10201-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------################################################## CGAGACGCTGCCGTCGGATGGGACTGAAGACGAAGCTGATGCCAACACAAGTAAGCACAAGTACTTTATGAATAAAAGCAATTACTAATGTATTTTCTACTCGCTCTCTGTCATAGCCGGAGTGCGTCGCTCCAGACGCGGCCAGGTGCAGCTGTGCAACACTTGG **************************************************...............((((((.....(((((.(((....))).)).......))).....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060673 9_ovaries_total |
SRR1106728 larvae |
V116 male body |
SRR060681 Argx9_testes_total |
SRR060670 9_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......GCTGCCGTCGGATGGGACTGAAG......................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GATGGGACTGAGGAC....................................................................................................................................... | 15 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................GATGGGATTGAAGACAAGGC.................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................GATGCCATTGAAGACGAAGCT................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................TAATGTATGTTCTCCTCGGT............................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................ATGTGGTTGCTACTCGCTCT........................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................CTGAAGACGAAGCTAACGAC........................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............GCCAGATGGGACTGAGGAC....................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCTCTGCGACGGCAGCCTACCCTGACTTCTGCTTCGACTACGGTTGTGTTCATTCGTGTTCATGAAATACTTATTTTCGTTAATGATTACATAAAAGATGAGCGAGAGACAGTATCGGCCTCACGCAGCGAGGTCTGCGCCGGTCCACGTCGACACGTTGTGAACC
**************************************************...............((((((.....(((((.(((....))).)).......))).....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060662 9x160_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
V053 head |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
M061 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................GCGCCGGTCCATGTCGAAA........... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AAGATGAGGGAGACACAG...................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AAGATGAGGGAGATACAG...................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................ATAAGATGAGGGAGATACAG...................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................TAAAGGATGAACGAGAGA......................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................CGCGGGCTGCGACGGTCCA................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................TAAGATGAGGGAGATACAG...................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:6925293-6925458 - | dvi_4245 | CGAGA-CGC--------------TGCCGTCGGATG------GGA-----------CTGAAG---------ACGAAGCTGATGCCAACACAAGT----AAGCACAAGTA-CT-T---TATGAATAAAAGCAATTACTAATGTATTTTCTACTCGCT-C-TCTG--T-C-ATAGCCGGAGTGCGTCGCTCCAGACGCGGCCAGGTGCAGCTGTGCAACACTTGG |
| droMoj3 | scaffold_6540:25687169-25687322 + | TGCCA-CTC--------------TTCCGCCGGATGAGCGAGATGACG-----------ATGC---CA------------------ACACGAGT----AAGTAAACTGCTGGCAAATGAC---TAGGCCAATCCACTAATGCATACTCAATTT--------------CATCAGCTGGCGTTCGTCGTTCCAGACGTGGTCAGGTGCCGCTGTGCAATACCTGG | |
| droGri2 | scaffold_15074:4867214-4867379 - | CGAGA-CGC--------------TGCCGCCGGAGG------AGA-----------ATGAAGCACGCAATGCCCACGATG------AGACAAGT----AAGAAAATGTA-TT-T---CAATTGCTA----TCAACGAATTA--TGTTTATATTTTT-CC-ATGCAA-AACCAGGTGGCGTGCGCAGATCGAGACGTGGTCAAGTGCCGTTGCGCAACACTTGG | |
| droWil2 | scf2_1100000004909:7015507-7015568 - | CAATGAT---------------A------------AGGATGATAGTG-----------ATAAGAATAATGATGATGATGATGCCAACAATAAT----AAACA----------------------------------------------------------------------------------------------------------------------CA | |
| dp5 | 2:17288268-17288427 - | AAGGAACTCCGTCATACATAATGAA------GTGGAAGTGGATT--------------TCAA---TGAA------------------TCTTGT----AAGTAGTACCA-----AACGATATGTTA----TTGCAATATTT-TCATTCAATTTT-TTCG---G--T-TATTAGCAGCCATTCGGCGGTCTCGGCGTGGTCAGGTACCATTAAAGAACACTTGG | |
| droPer2 | scaffold_0:11133061-11133219 - | AGGGA-CTCCGTGATATATAATGAA------GTGGAAGTGGATT--------------TCAA---TGAA------------------TCTTGT----AAGTAGTACCA-----AACGATATGATA----TTGCAATATTT-TCATTCAATTTT-TTCG---G--T-TATTCGCAGCCCTTCGGCGGTCCCGGCGTGGTCAGGTACCATTTAAGAACACTTGG | |
| droFic1 | scf7180000454106:2761283-2761444 - | ATTGAAGCTTTCCAAAATTCCAGTG------GAGACGGAGGATCAGG-----------ACCA---CGAT------------------ACCTGT----GAGTATATTTT-----AACAACGACTTA----ATTAAAACTTA-ATATTAACATTA-TTTC---G--T-T-GTAGCCGGCATTCGGCGCTCGAAGCGAGGCCAAAAACCACTCCAAATGAGTTGG | |
| droEle1 | scf7180000491212:1416165-1416310 - | TGGA-----------------------------AATGGATGAAAACG-----------ACCA---CAAC------------------ACCTGTGAGTAACTAAAAAAA-----AACTATGATTTC----TTGAAATATTT-TATTTAT-ATTATT-TT-ATTG-T-TCTTAGCCGGCATTCGGCGTTCGAAGCGAGGCCATGTGCCACTCCAGAATACCTGG | |
| droRho1 | scf7180000780121:12713-12862 + | TCCGG------------------TG------GTAATGGATGAAGGCG-----------ACCA---CAAT------------------ACCTGT----GAGTATATTTTGAAATAACGTT---TTAACCTTCCATTAAACC--TATTTTTGTTT-----TCTGT-T-ACTCAGCCGGCATTCGGCGTTCAAAACGAGGCCAAGTGCCAATCCAGATGACTTTT | |
| droTak1 | scf7180000414445:283583-283601 + | CGAGGTT---------------A---------------------ACG-----------ATGT---CA------------------TC--------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 2R:12081074-12081121 + | ACATGGT---------------G---------------------GGGTGGCCGCCCTGAAG-----------------------------------------------------------------------------------------------------------------------CAGCAACTGCTGCAAAAGCAGGT------------------- | |
| droEre2 | scaffold_4770:17181684-17181824 - | AATTC-CGG--------------TGGAATCAGAGG------AA-----------------CT---AAAT------------------ACAAGT----AAGTATATTTTATATTAGGTGC---TTTA-------TTTTTTA--TATTATTATTT-----TCTGT-TTCTTCAGCTGGTATTCGGCGATCGAAGCGAGGTCAAGTTCCACTCCAGATGAGTTGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:35 PM