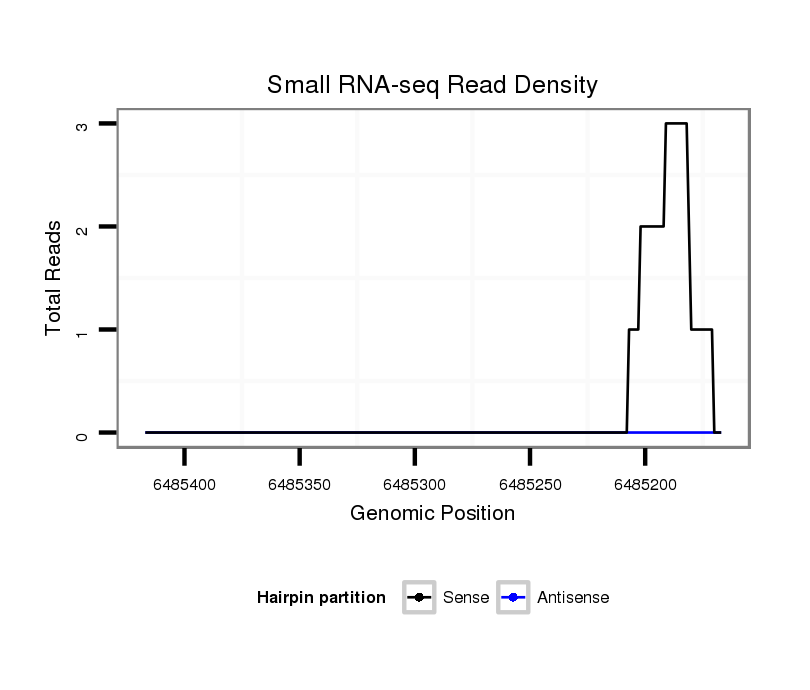
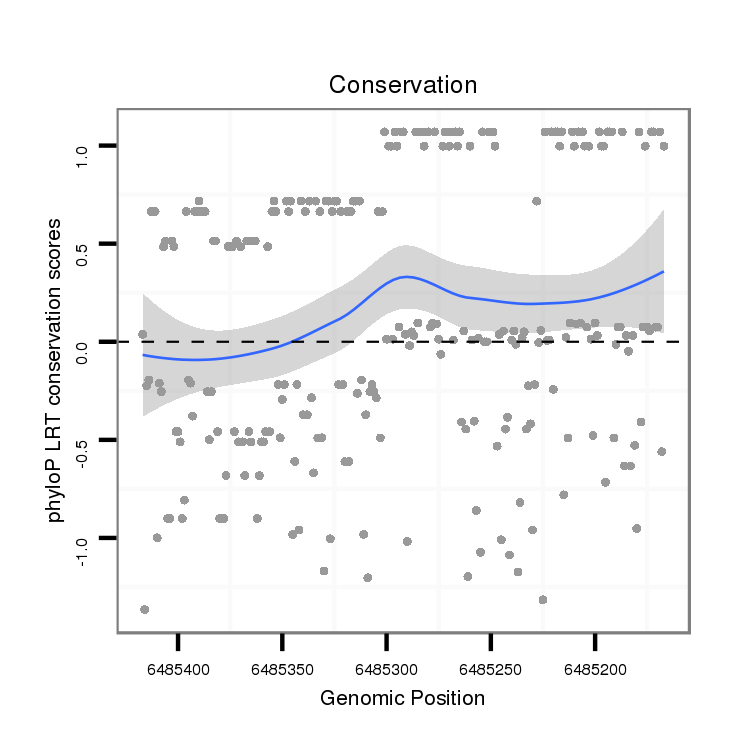
ID:dvi_4183 |
Coordinate:scaffold_12855:6485217-6485367 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ10224-cds]; exon [dvir_GLEANR_10163:3]; intron [Dvir\GJ10224-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGGCCGCCATGAGCAGGATGGCGAACGACCCAATTATGCCAGGGGATCTGTTTTACGTTTGTCAGACCATGACCATAGGCATCATCATAAAGATGCCGCCGATATGCGCAGTTGTGAGCCCAGCTTTACAAAATTCAAGATACCCTAGTGAGTAATCGGGCAGTTGTAAGCGATAGACTACCTTAGATATCCTTCGTATAGTGAGATGATATGCGCCAAACGGTATCAGGATTCAATCAAGGCTTACGAGG **************************************************...((((...((((.....))))....((((((........)))))).(((((((.(((((((((((((...)).)))))).............)).))).)).)))))........(((.((((..(((...))).)))))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060681 Argx9_testes_total |
M027 male body |
V053 head |
SRR060687 9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060683 160_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................CCAAACGGTATCAGGATTCAAT.............. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CAGGATTCAATCAAGGCTTAC.... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................ATGCGCCAAACGGTATCAGGATTCAA............... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........AGCAGGATGGCGAACGCGCCT........................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TCCTTCGTAGAGTGATGT............................................ | 18 | 3 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 1 |
| ..................................................................................................................................................................................................CGTGTCGTGAGATGTTATG...................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................CGTCGAGCTGCGCAGTTGTG....................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................CGGTATGCGGAGTTGTAA...................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TCCGGCGGTACTCGTCCTACCGCTTGCTGGGTTAATACGGTCCCCTAGACAAAATGCAAACAGTCTGGTACTGGTATCCGTAGTAGTATTTCTACGGCGGCTATACGCGTCAACACTCGGGTCGAAATGTTTTAAGTTCTATGGGATCACTCATTAGCCCGTCAACATTCGCTATCTGATGGAATCTATAGGAAGCATATCACTCTACTATACGCGGTTTGCCATAGTCCTAAGTTAGTTCCGAATGCTCC
**************************************************...((((...((((.....))))....((((((........)))))).(((((((.(((((((((((((...)).)))))).............)).))).)).)))))........(((.((((..(((...))).)))))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060683 160_testes_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................CGGGGCGAAATCTTTTAA.................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TGCCAAAGCCCTAAGGTAG............. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................TAGAGGAAGCATTTCACTT.............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................GTTTTAAGTGCTATTGGGT........................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:6485167-6485417 - | dvi_4183 | AGGCCGCCATGAGCAGGATGGCGAACGACCCAATTATGCCAGGGGATCTGTTTTACGTTTGTCAGACC--------------------------ATGACCATAGGCATCATCATAAAGATGCCGCCGATATGCGCAGTTGTGAGCCCAGCTTTACAAAATTCAAGATACCCTAGTGAGTAAT-----CG-------GGCAGTTGTAAGCGATA----GACTACCTTAG----ATATCCTTCGTAT-AGTGAGATGATATGCGCCAAACGGTATCAGGATTCAATCAAGGCTTACGAGG |
| droMoj3 | scaffold_6540:6851980-6852229 - | GCGCCGCAAC----------CCGAACGACCCGCC----------------------------CAGACCTAAAGTCGAGATGGATATGATTTTATATGATGAAAAGTACAAGCCCAACGATACTCCGGATTTGTATGACCGGGAGCCCAGCTTCCGGAAATTCAAGATACCCTAGTAAGTGTAAGTATAG-------AAATCT--TAAGAGCATCGCGAGTAATATCTGTTCATAATATCTCATAT-AGTCCGATGATATGCGCCAAACGATATCAGGACTCGATTAGGGCATACGAAG | |
| droGri2 | scaffold_14906:7618534-7618783 + | GAATCGCTGTGACGAGAGACCCAGCCGACCCAATTACCGGCGGGAAACATTCATAGTAACGCCAGTGA--------------------------TTGAACTCAAGTACAAGCCGAATGATGCCGCCGATATAAAGAGTCGGGAGCCCAGCTTCGCAAAATTCAAGATACCCTAGTAAGTGAT-----GG-------AAATGT--TAAGCATTA----GCTTCGACTAA----TCTTACGTAATTTAAGTGAAATGATATGCGCAAAACGGTATTCGGACTGGAAAAGGGCTTACGAGG | |
| droWil2 | scf2_1100000004902:7463613-7463760 - | GCA-------------------------------------------------------------------------------------------------------------------------------------------ATCCAAGTTTCGCACAGTTCAAAGTTAGCTAGTAAGTAAATAACTAGGCCACTAAGTTCT--TAAGTGATTTATCAATTATCTTTA--------ACATCATTT-AGTAATGTGTTGTGTGATGCACGCTATAAAAAAGCTCGAAAAGTTTATAATG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/16/2015 at 08:09 PM