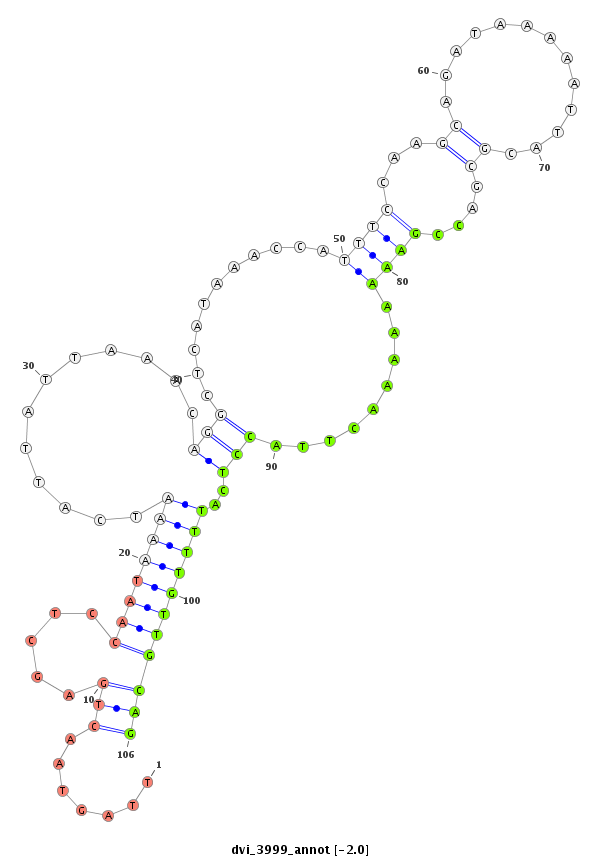
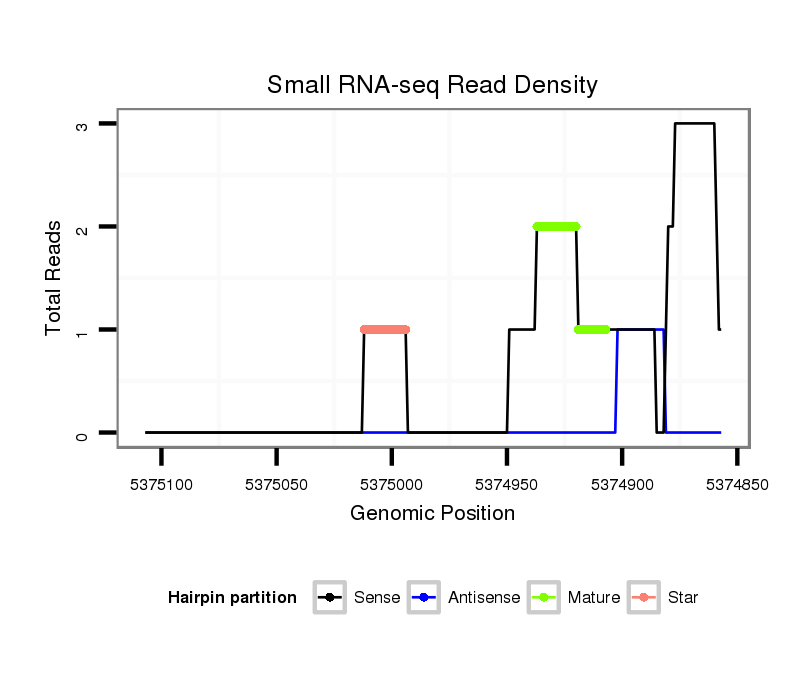
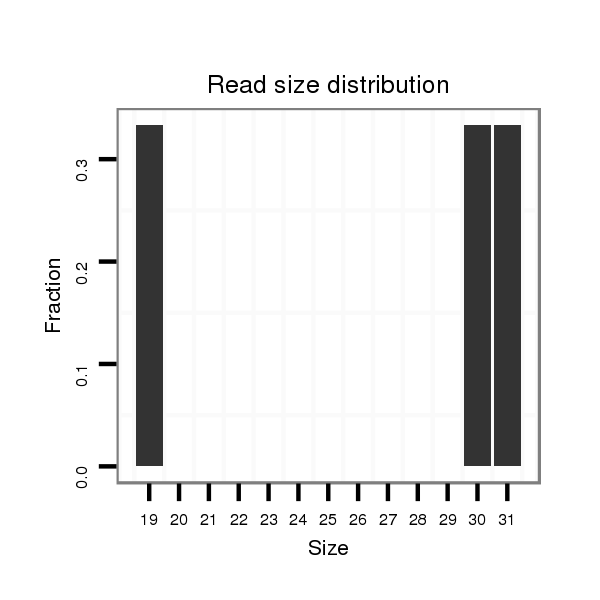
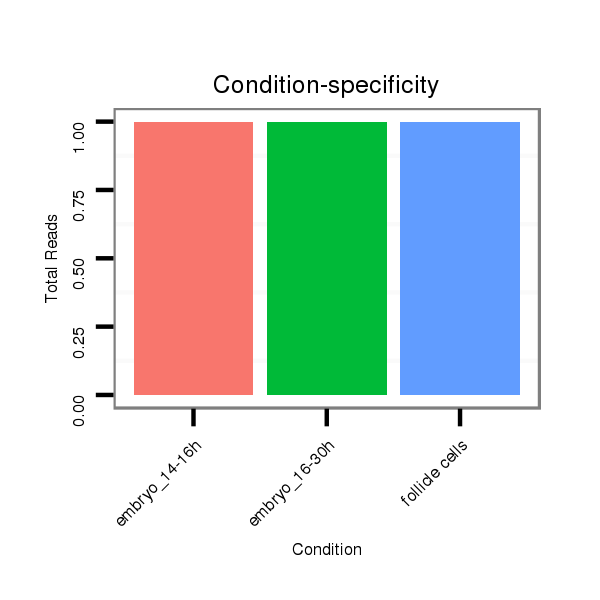
| ACCAATTAAT---------G-------------------------CCG---CATCAA------------------------------------------------------------------------------------------------------------------------CAACAA-----CAAAAACAG-----------------------------------------------------------------------------------------------------------------------------------------------A-----------------------------------------------------------------------------------------------------AGTAGCCGCCCACAACAGCCAT----AGCCTGTAACACAAATAC-----------------------CACCC-------------------------------------------------------------------------------------------------------------------------------------------------------------AGAAA----------------------------------------------GCACCCCATCA----------AGCT------------------GCA-----GCATCGTCTA------CAG---------CCATAATAACAT----CA---ACTACAACATCGTCTG-TGGCTGC---------AGCCCCGG-----C | Size | Hit Count | Total Norm | Total | M021
Embryo | M042
Female-body | V042
Embryo | V050
Head | V057
Head | V111
Male-body |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGG........................... | 25 | 5 | 7.20 | 36 | 0 | 36 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-T............................. | 23 | 5 | 4.40 | 22 | 0 | 18 | 3 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGGC.......................... | 26 | 5 | 4.20 | 21 | 7 | 14 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TG............................ | 24 | 5 | 3.00 | 15 | 2 | 13 | 0 | 0 | 0 | 0 |
| .....................................................TCAA------------------------------------------------------------------------------------------------------------------------CAACAA-----CAAAAACAG-----------------------------------------------------------------------------------------------------------------------------------------------A-----------------------------------------------------------------------------------------------------AGTAG...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-.............................. | 22 | 5 | 2.80 | 14 | 3 | 9 | 0 | 1 | 0 | 1 |
| .....................................................TCAA------------------------------------------------------------------------------------------------------------------------CAACAA-----CAAAAACAG-----------------------------------------------------------------------------------------------------------------------------------------------A-----------------------------------------------------------------------------------------------------AGTA....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCT................................ | 21 | 5 | 1.40 | 7 | 4 | 2 | 1 | 0 | 0 | 0 |
| .....................................................TCAA------------------------------------------------------------------------------------------------------------------------CAACAA-----CAAAAACAG-----------------------------------------------------------------------------------------------------------------------------------------------A-----------------------------------------------------------------------------------------------------AGTAGC..................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGGCTG........................ | 28 | 5 | 0.60 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAT----CA---ACTACAACATCGTCTG-TGGC.......................... | 25 | 5 | 0.60 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T------------------GCA-----GCATCGTCTA------CAG---------CCAT................................................................ | 21 | 6 | 0.50 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGA........................... | 25 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................TCAA------------------------------------------------------------------------------------------------------------------------CAACAA-----CAAAAACAG-----------------------------------------------------------------------------------------------------------------------------------------------A-----------------------------------------------------------------------------------------------------AGTAGCA.................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-G............................. | 23 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAT----CA---ACTACAACATCGTCTG-TGG........................... | 24 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A---ACTACAACATCGTCTG-T............................. | 18 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGT........................... | 25 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGGCTGG---------.............. | 29 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT----CA---ACTACAACATCGTCTG-TGAAA......................... | 27 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T----CA---ACTACAACATCGTCTG-TGGC.......................... | 23 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAT----CA---ACTACAACATCGTCTG-T............................. | 22 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAT----CA---ACTACAACATCGTCTG-.............................. | 21 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T------------------GCA-----GCATCGTCTA------CAG---------CCA................................................................. | 20 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A-----GCATCGTCTA------CAG---------CCATA............................................................... | 19 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATCGTCTA------CAG---------CCATAATA............................................................ | 19 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T------------------GCA-----GCATCGTCTA------CAG---------CCATA............................................................... | 22 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |