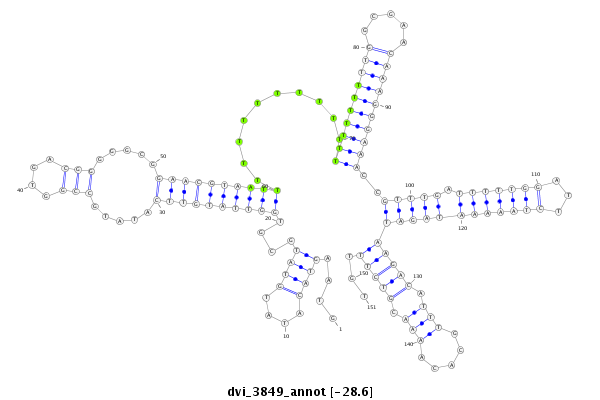
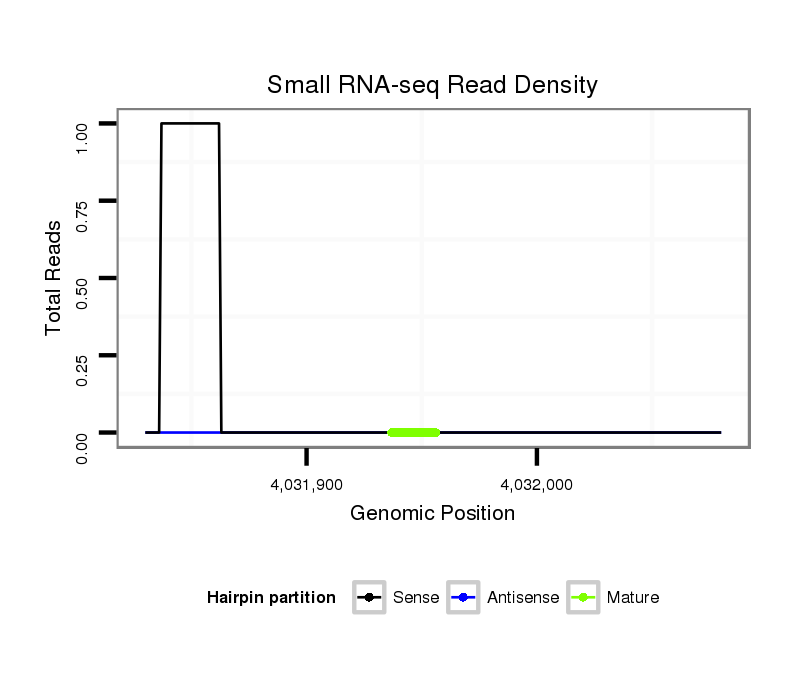
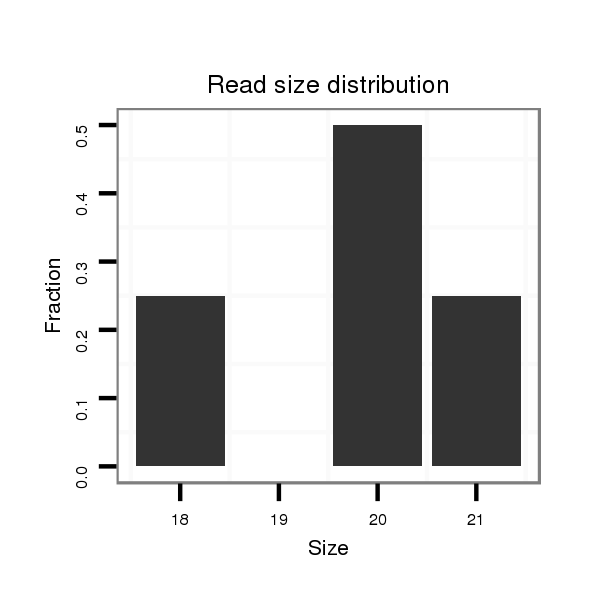
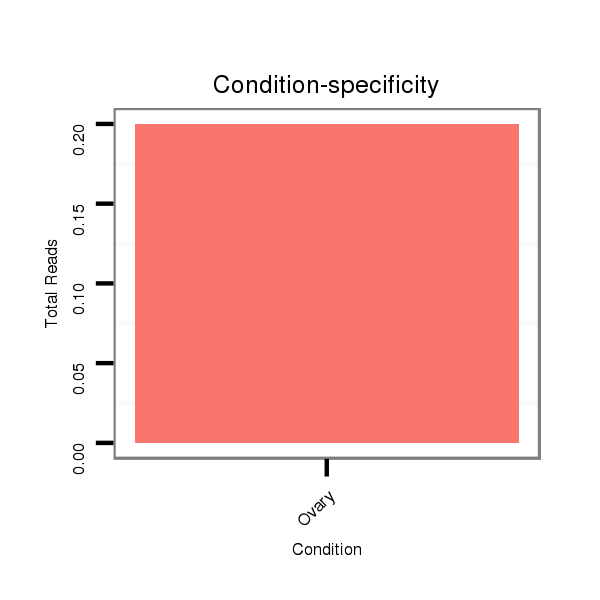
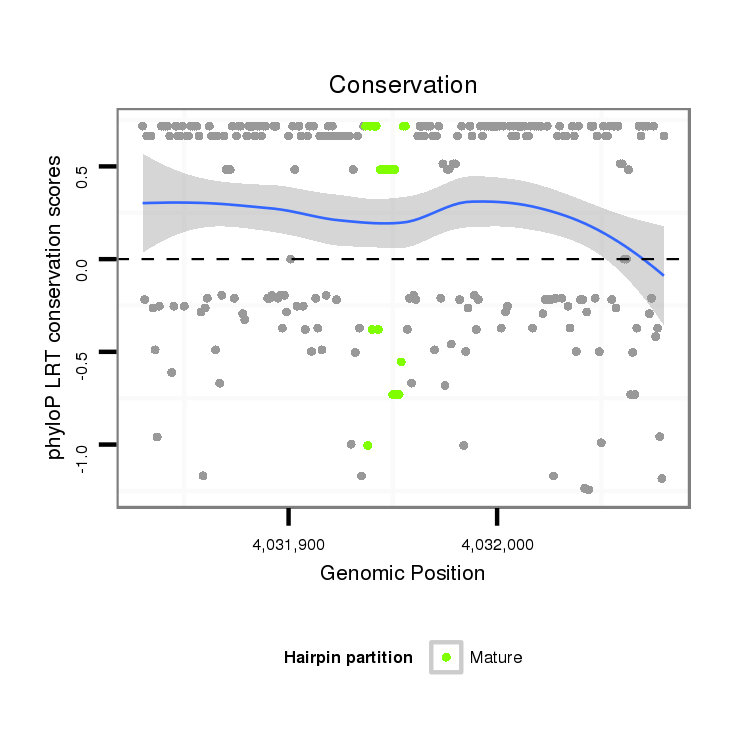
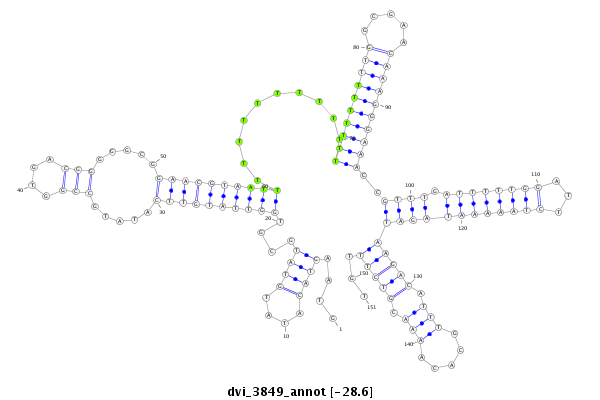
ID:dvi_3849 |
Coordinate:scaffold_12855:4031880-4032030 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -28.6 | -28.5 | -28.5 |
 |
 |
 |
exon [dvir_GLEANR_10708:1]; CDS [Dvir\GJ10808-cds]; intron [Dvir\GJ10808-in]
| Name | Class | Family | Strand |
| (T)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TACGCACTATATTCCATCGTAGCTATAGTAATAGCACGCCAATTAGAACGGTAAGTACATATGTATGCGTGGTTATGTTCATATGCCGGTGACGGGGGCGGAACGTAATTTTTTTTTTTTTTTTTTTTTGGCGAACAAAGGGAAACCGTTTGATTTTTGGATTCTAAAAATAGATAAGACATTTGCACAAAACGTCTTTGTTGCTGTCTAAACTCTTTGAATGCACAAATATATGCACACATACATTGTTG **************************************************....((((....))))....((((((((((.....(((....))).....))))))))))..........((((((((((.....))))))))))..(((((.(((((((...))))))))))))(((((.(((.....))).)))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060669 160x9_females_carcasses_total |
M047 female body |
SRR060670 9_testes_total |
SRR060680 9xArg_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
M028 head |
V053 head |
SRR060654 160x9_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......TATATTCCATCGTAGCTATAGTAATA.......................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAAGTACATATGTATGTGTGG................................................................................................................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TGGCGAACAAAGCGACACGGT...................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................ATTCCGGTGACGGAGGCGG...................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TTCAGATGCCGCTGAGGGGGGCG....................................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ATTTTTTTTTTTTTTTTTTT............................................................................................................................ | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................ACAAAACTTCTTCGTTGC............................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................ATGATTTTTGGATTGTAAAA.................................................................................. | 20 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................ATTTTTTTTTTTTTTTTTTTT........................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................TTTTGGATTCTAGAAAAA............................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................ATTTTTTTTTTTTTTTTT.............................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ATGCGTGATATAAGGTAGCATCGATATCATTATCGTGCGGTTAATCTTGCCATTCATGTATACATACGCACCAATACAAGTATACGGCCACTGCCCCCGCCTTGCATTAAAAAAAAAAAAAAAAAAAAACCGCTTGTTTCCCTTTGGCAAACTAAAAACCTAAGATTTTTATCTATTCTGTAAACGTGTTTTGCAGAAACAACGACAGATTTGAGAAACTTACGTGTTTATATACGTGTGTATGTAACAAC
**************************************************....((((....))))....((((((((((.....(((....))).....))))))))))..........((((((((((.....))))))))))..(((((.(((((((...))))))))))))(((((.(((.....))).)))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060685 9xArg_0-2h_embryos_total |
SRR060688 160_ovaries_total |
M027 male body |
V047 embryo |
M047 female body |
M028 head |
SRR060675 140x9_ovaries_total |
SRR060676 9xArg_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................GATAGATATCCTTATCGTGC..................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................AAAAAACTGCTTGTTGTCCTTT.......................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CAGATGTGAGAAGCTTCCG........................... | 19 | 3 | 12 | 0.17 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| AGGCATGATATGAGGTAGC........................................................................................................................................................................................................................................ | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................CCGCTTGTCTCCCTTTTG........................................................................................................ | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................AAAAAAAAAAAAACAGCTTGT.................................................................................................................. | 21 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AAAAAAACAACCGCTTATTT................................................................................................................ | 20 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................AAAAAAAAAAAAAAACCGTCTGTTT................................................................................................................ | 25 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................AAAAAAAAAAAAGCGCTGGTT................................................................................................................. | 21 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................TAAAAAAAAAAAAAAAAAAA............................................................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:4031830-4032080 + | dvi_3849 | TACGCACTATATTCCATCGTAGCTATAGTAATAGCACGCCAATTAGAACGGTAAGTACATATGTATGCGTGGTTATGT----------------------TCATATGCCG---GTGACGGGGGCGGAAC--GTAATTTTTTTTTTTTTTTTTTTTTGGCGAACAAAGGGAA--ACCGTTTGATTTTTGGATTCTAAAAATAGATAAGACATTTGCACAAAA--CGTCTTTGTTGCTGTCTAAACTCTTTGAATGCACAAATATATGCACACATACATTGTTG |
| droMoj3 | scaffold_6540:8346596-8346840 + | TACGCTCAGTATTCGGTCGTGGCTATAGCGTTAGCCCTCCAATTAGAACTGTAAGTACATATGTATGTGCG-TTGTGC----------------------TCAGATACCG---GTGACAGGGGCGTAGCAAACAAGTTTTTTTTTTTGTGGATTTTTGTGAACAAAGGGAATA-------GATGGATGGATTCTAAAAATAAACGAGACATTTGCATAAAAATCATTTCTGTTGCCATCGAAAGCATTTGCTTGCATATA----TTTCTACATACATAACGG | |
| droGri2 | scaffold_14906:6655096-6655351 + | TTCGCAGCATATTCCATCGTAGCTATAGCCACAGCCCTTC---TGGAAAGGTAAGTACATGCATACATACG-T-ATGTATATTACTAAGAAATATACATTGCATACACGGATGATGACGGGGGCGA--CAAAGAAATGTTG-----------TTGCTACGAACAAAGCGAGCGAACGCTTGTTATTTGACATCTAAAAATAAACAAGACATTTGCATAAAA--AGTCAGCGTCGCTATCTATTTCGTTCGACTGCACAAATA------TACATAAGTAAACG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 11:28 PM