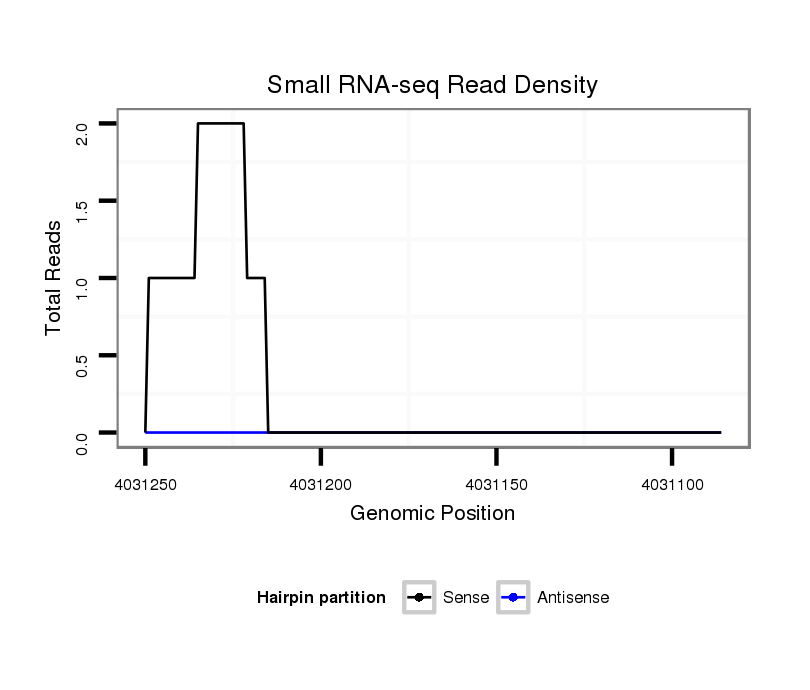
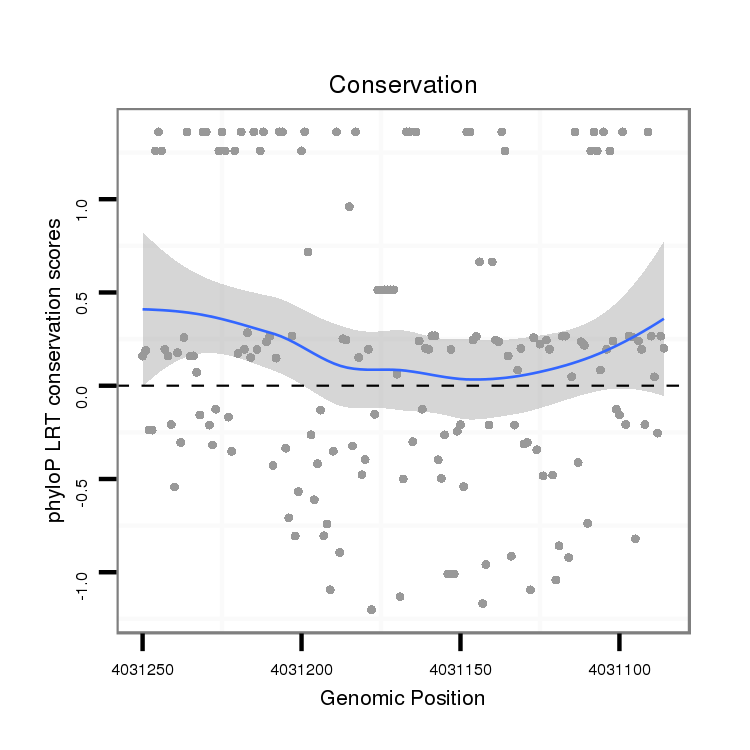
ID:dvi_3848 |
Coordinate:scaffold_12855:4031136-4031200 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ10343-cds]; exon [dvir_GLEANR_10273:1]; exon [dvir_GLEANR_10273:2]; CDS [Dvir\GJ10343-cds]; intron [Dvir\GJ10343-in]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------################################################## AGAGGAGAATGCCATAGCCAAACAGAGCGCCAAATTTCACAGCAAATTTGGTATGAATGCCTGCTTTTAAGAGTTTAAATATAAACTAAAAATATTCTGATTTTTTCATACACAGCAACAACCATGGAAAATATTAAACGACAGCAACATGAATCGAAAAAATTA **************************************************(((((((..........((..((((((....)))))).....))..........)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060672 9x160_females_carcasses_total |
V053 head |
V047 embryo |
SRR060669 160x9_females_carcasses_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............AGCCAAACAGAGCGCCAAAT.................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .GAGGAGAATGCCATAGCCAAACAGAGCG........................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................GTATTAAACGACAGCAACATGAAT........... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TATGACTGCCTGCTTTGAGGAG............................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CCATCGAAAAGTTTAAACGA........................ | 20 | 3 | 17 | 0.24 | 4 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| ..............................AATATTTCACAGCAAAATTGG.................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCTCCTCTTACGGTATCGGTTTGTCTCGCGGTTTAAAGTGTCGTTTAAACCATACTTACGGACGAAAATTCTCAAATTTATATTTGATTTTTATAAGACTAAAAAAGTATGTGTCGTTGTTGGTACCTTTTATAATTTGCTGTCGTTGTACTTAGCTTTTTTAAT
**************************************************(((((((..........((..((((((....)))))).....))..........)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V053 head |
V047 embryo |
M061 embryo |
GSM1528803 follicle cells |
SRR060677 Argx9_ovaries_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................CTGTCCTTGTACATAGCTTTGT.... | 22 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CTGTCCTTGTACATAGCTT....... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................TGTCCTTGTACATAGCTTT...... | 19 | 2 | 3 | 0.67 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................TGTCCTTGTACATAGCTTGT..... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................AAAAAGTTTGAGTCGTTGT............................................. | 19 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................CGGGCTGTCTCTCGGTTTAA................................................................................................................................. | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................CGGACGAAACTTGTCAGAT........................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:4031086-4031250 - | dvi_3848 | AGAGGAGAATGCCATAGCCAAACAGAGCGCCAAATTTCACAGCAAATTTGGT----ATGAAT----------GCCTGC---TTTTAAGAGTTTAAATATAAACTAAAAATATT--CTGAT--TTT-TTCATACACAGCAACAACCATGGAAAATATTAAACGACAGCAACATGAATCGAAAAAA-TTA--- |
| droMoj3 | scaffold_6540:8345841-8346000 - | ACAGGAGAATGCACTAGCTAAGCAGAGCGCCAAGTTTCACAGCAAAATCGGT----AACTAA----------AATTGT---T--TACAAC-------TGGAATTGAAATATGAATTTAATATTTT-TTCGAACACAGCGGCAAAACATCAACTTATAAAACGCCAGCAACGTAAATTGGAAAAA-TTA--- | |
| droGri2 | scaffold_14906:6654340-6654511 - | ACTGGAGAATACTATAGCTAAGAAGAGTACGAAATTTCACACCAACTCCGGT----ATGTAAATGAA------TCTTC---GTCTACAAGCTTAAATAACAATTGAAATAATT--ATTGTTTGTTTGTCCCGCCCAGCGGCACTTATGACAACGATTGACCTCCAGCAACGTAAATTCGAAAAG-CTA--- | |
| dp5 | 2:11387716-11387884 - | CCGCGAGGCGTCCATCAGCAAACGGAGCTCCAGAGTCCACAAAAATATTCGTGAGT----CCCGGCAAGCGTTCATCCTATT--TCAGGC-------ATCAATTGGTGTAC--ATTCAACGTGTT-TT-----ACAGTTAGTGCCAAGGAGGCCATCGAACGGCAGGAGCGCGACTTTGGCAAG-CTGCAA | |
| droPer2 | scaffold_0:9126014-9126182 + | CCGCGAGGCGTCCATCAGCAAACGGAGCTCCAGAGTCCACAAAAATATTAGTGAGT----CCCGGCAAGCGTTCATCCTATT--TCAGGC-------ATCAATTGGTGTAC--ATTCAACGTGTT-TT-----ACAGTTAGTGCCAAGGAGGCCATCGAACGGCAGGAGCGCGACTTTGGCAAG-CTGCAA | |
| droAna3 | scaffold_13340:9387264-9387427 - | CCGTGAGACCAACATCAGCAAAATGAGTTCCAAATTTCATGACAATCTGCGTTAGT----TT--GCAAACAAAACTAC---TTTTAAAAAC-------CCAAATGTAATAC--AAATTTGTTATT-TA-----ATAGTCACAGCAATGGAAAAAGCGGACTGGCAGCAACGAGAGCTCGAGAGGATCG--- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
Generated: 05/16/2015 at 11:19 PM