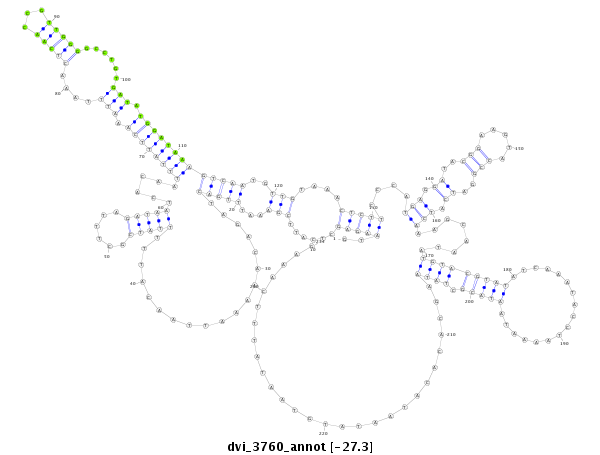
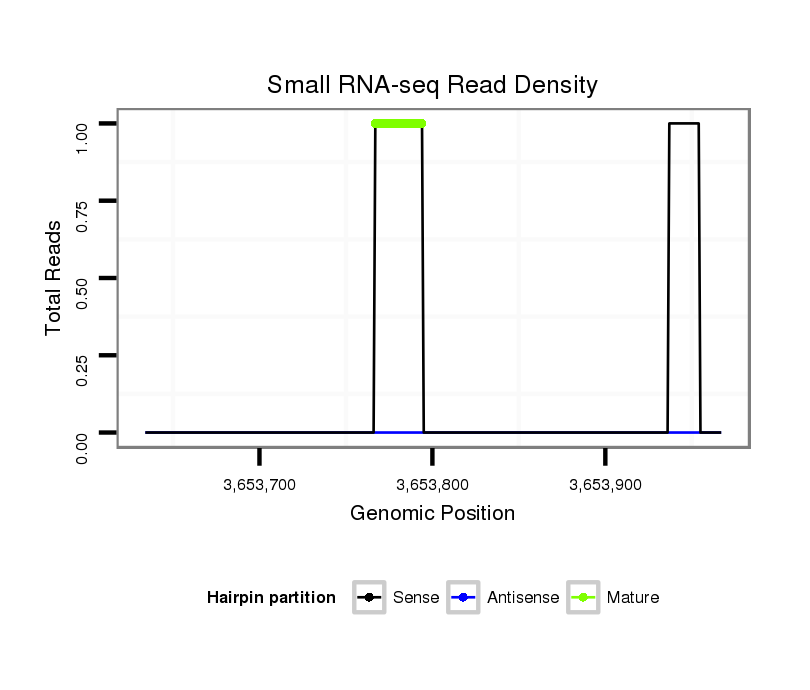
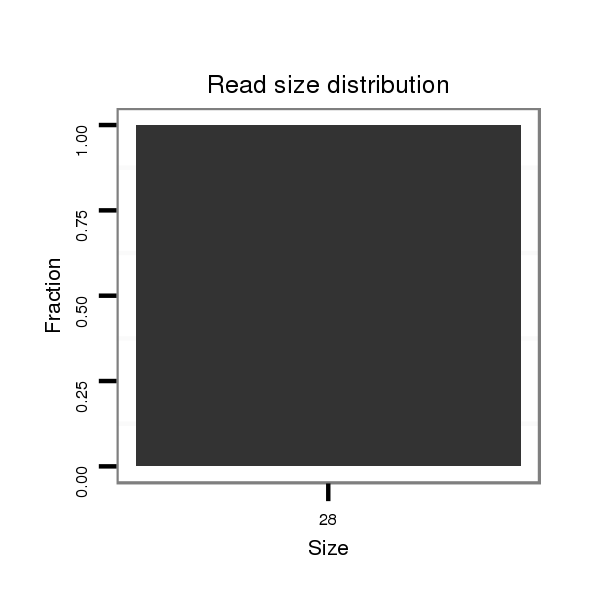
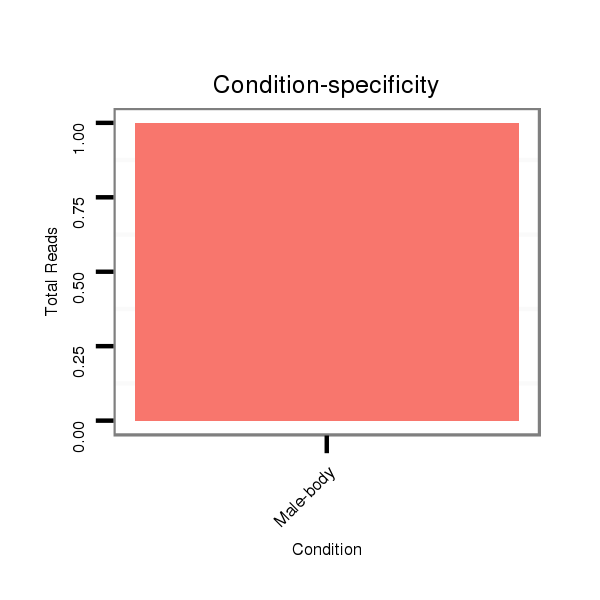
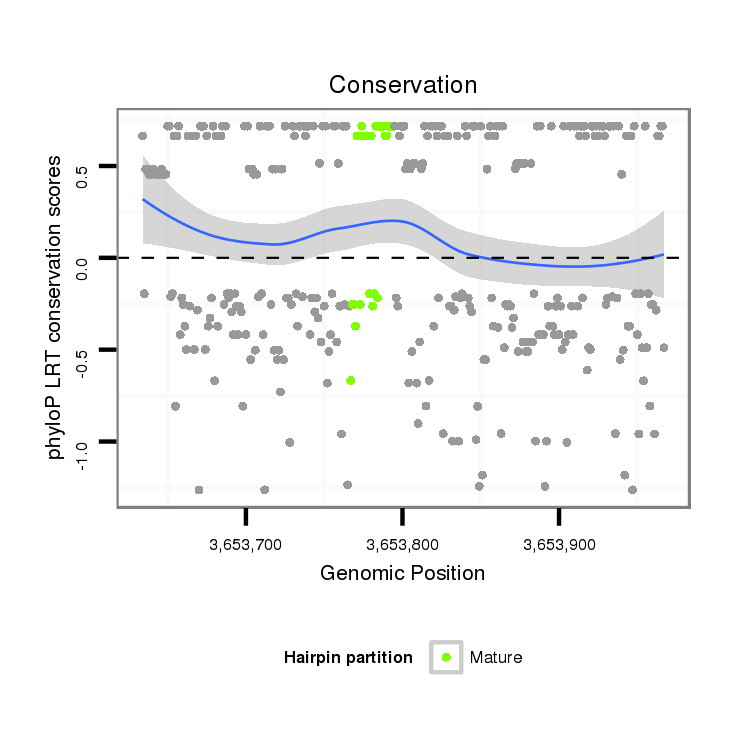
ID:dvi_3760 |
Coordinate:scaffold_12855:3653684-3653917 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.28 | -27.1 | -27.1 |
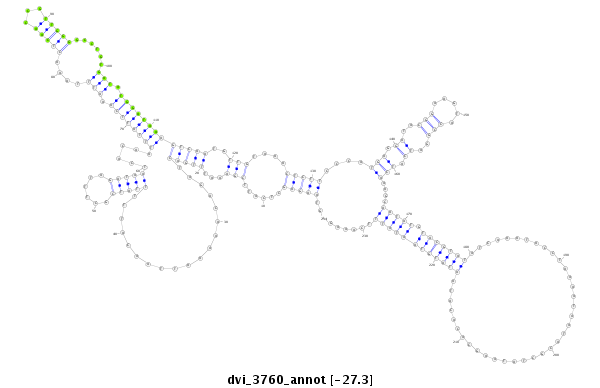 |
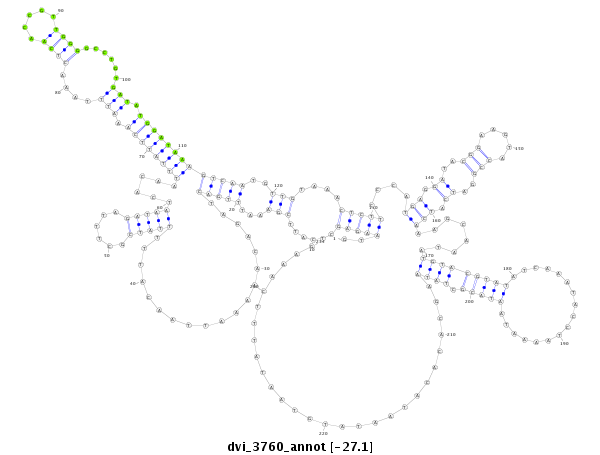 |
 |
CDS [Dvir\GJ10787-cds]; CDS [Dvir\GJ10787-cds]; exon [dvir_GLEANR_10688:3]; exon [dvir_GLEANR_10688:4]; intron [Dvir\GJ10787-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CGATCCCAGCGGTCCGAATGCGGAAAACAGAGGAGTCAAAAGGCGAGCGGGTAAGAGCTCATTCGAAATTTGACTAGACAAAAATTAACATTTTTATCGCTTTAGATAATCACAATTTATTCAAATTTAAACTCAACCGTTGGGGCCTGTGATATGGATAAAGTCAATGTTGTAAACTCTTCCCATGAGGATACGGAAGTACCGGATCATCAAAGCAATATGTACGTATATCAAATACCTAAAATAATACGCTATAAGCACACATAATATGTAATATTTCAAAGCGAATGTGATTGTGATAAAGATGGAGGCGCAGTACAGTGCCATAACCAAG **************************************************..(((((......(((...(((((...................(((((......)))))......((((((((.(((....(((((...)))))......))).)))))))))))))..)))....)))))....(((.((..(((.....)))..)).)))........(((((((((.................))))).))))............................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M027 male body |
SRR060659 Argentina_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060678 9x140_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
V116 male body |
SRR060661 160x9_0-2h_embryos_total |
M047 female body |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................GATGGGGAAGTACCGGTTCA............................................................................................................................. | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CCTAGGGGTCCGAATGGGGAA..................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................ATCGAATGTGATTGTGATAAAGAT............................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CAACCGTTGGGGCCTGTGATATGGATAA............................................................................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................GATGGAGGCGCAGTACAG............. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................TGGGATCTTGATAAAGATGGAG........................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................TGTGCTGGTGAGAAAGATGGAG........................ | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................TGATTCTGATAAAGATGACGGC...................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................TGATTGTGAGAAAGAGGGAGC....................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................GGTAAGAACTCATTGGAA........................................................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................GGTGGAGGCGTAGTACACT............ | 19 | 3 | 16 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| ..........................................................................................................................................GTTGGGACCGGTGGTATGGA................................................................................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................GATAAAGTCAATGCTGAA................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCTAGGGTCGCCAGGCTTACGCCTTTTGTCTCCTCAGTTTTCCGCTCGCCCATTCTCGAGTAAGCTTTAAACTGATCTGTTTTTAATTGTAAAAATAGCGAAATCTATTAGTGTTAAATAAGTTTAAATTTGAGTTGGCAACCCCGGACACTATACCTATTTCAGTTACAACATTTGAGAAGGGTACTCCTATGCCTTCATGGCCTAGTAGTTTCGTTATACATGCATATAGTTTATGGATTTTATTATGCGATATTCGTGTGTATTATACATTATAAAGTTTCGCTTACACTAACACTATTTCTACCTCCGCGTCATGTCACGGTATTGGTTC
**************************************************..(((((......(((...(((((...................(((((......)))))......((((((((.(((....(((((...)))))......))).)))))))))))))..)))....)))))....(((.((..(((.....)))..)).)))........(((((((((.................))))).))))............................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
M047 female body |
SRR060665 9_females_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060671 9x160_males_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
M027 male body |
SRR060659 Argentina_testes_total |
M028 head |
GSM1528803 follicle cells |
SRR060675 140x9_ovaries_total |
SRR060681 Argx9_testes_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................................................................................GTCGTGCCACGGAATTGGTT. | 20 | 3 | 2 | 18.00 | 36 | 8 | 4 | 2 | 3 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TCGTGCCACGGAATTGGTT. | 19 | 3 | 10 | 4.60 | 46 | 5 | 7 | 6 | 3 | 1 | 0 | 6 | 2 | 2 | 1 | 0 | 4 | 2 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................GTCGTGCCACGGAATTGGT.. | 19 | 3 | 7 | 1.43 | 10 | 1 | 1 | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................GTCGTGTCACGGAATTGGT.. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................ATGCCTTGGTGGCCTAGT............................................................................................................................. | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TATGCCTTGGTGGCCTAGTG............................................................................................................................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................ATGCCTTGGTGGCCTAGTG............................................................................................................................ | 19 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................GTCGTGCCACGGCATTGGTT. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GTATGCCTTGGTGGCCTAG.............................................................................................................................. | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TCGTGCCACGGAATTGGT.. | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ...........................................................................................................................................................................................................................................................................................................................CGTGCCACGGAATTGGTTC | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:3653634-3653967 + | dvi_3760 | CGATCCCAGCGGTCCG------AATGCGGAAAACAGAGGAGTCAAAAGGCGAGCGGGTAAGA-----GCTCATTCGAAATTTGACTAGACAAAA--------------------------------ATT-------AACATTTTTATCGCTTTAGATAATCACAATTTATTCAAATTTAAACTCAACCGTTGGGGCCTGT---GATATGGATAAAGTCAATGTTGTAAACTCTTCCCATGA---GGATACGGAAGTACCGGATCATCAAAGCAATATGTACGTATATCAAATACCTAAAATAATACGC-TATAAGCACACATAAT-ATGTAATATTTCAAAGCGAATGTGATTGTGATAAAGATGGAGGCGCAGTACAGTGCCATAACCAAG |
| droMoj3 | scaffold_6540:7947466-7947826 + | CGATCCCAGCGGTCCGCATAACAATGCCGATAGTCGTGGCGCGAAACGAAAATCAGGTGAGATAGTAGCACTAACCATATATGGCTAGTGAAAGTAGTCAGCAGTAGCAGTTTATGCCTTTCAAAAACTATCGATATGAAATTTGGTCGTTTTAGATTATCAA--TAT--ACAA--AAAGAGAAGGTCGCTGGGGCCAGTATTAATATGGATAAAAACAAT------------TGCAATAAAATGAATGCGGTTTCACAAGATCAACAATCAACATTGTATGTATGTCGTGAAA------------GAGTTTTTTATTATTAATATCT-GTAATTTACAAAGGGCATGTGATTGCAATAAGAAAGACGATGGAGATCCTTGCGGCTGCCAAG | |
| droGri2 | scaffold_14906:6310281-6310600 + | CA----------------CAACAACACCGATTATAGAGGAGCAAAAAGACGATCAGGTAAATAAAAAAAATAATACATA------CAATAAAAATAGTAACGAA-------------------------------------TAAAATCATTTCAGATTTTATCAGTTTCAATAAGTTCAAATTAAATCGTTGGGGTCTAT---GATATGGATAAAGTCAATGTGGAAACGCCTTGCAATAA---GGATTCGGAAACGCTGAGTCTTAAACCGAG---GTTTGTCTTTGAAATCCCTTAAGTTTCATATATTTTTCTATATTTTTT-ATAATATTTTTCATAGCCAATGTGATTGTGATGATGA---GGATGAAAAGTCTTCTGATGGCCAAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 09:32 PM