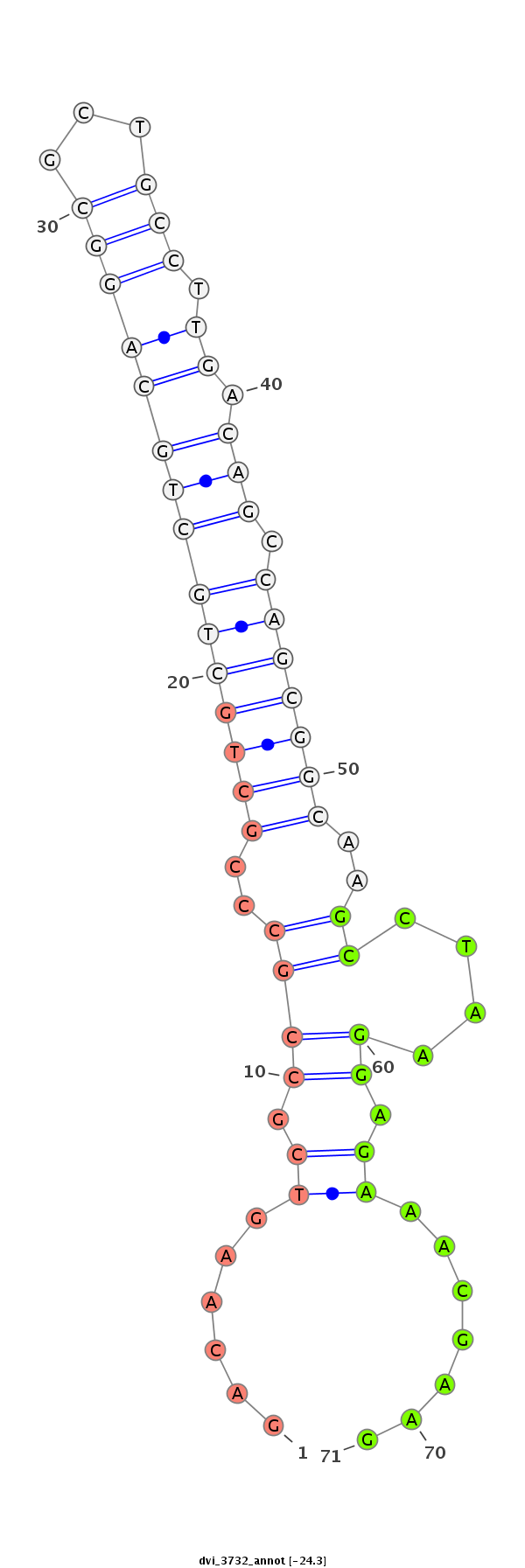
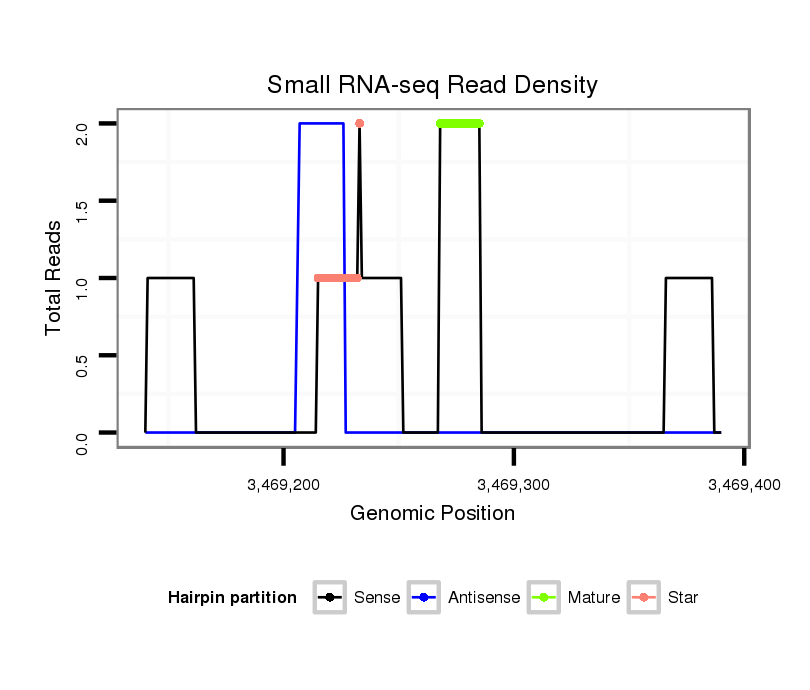
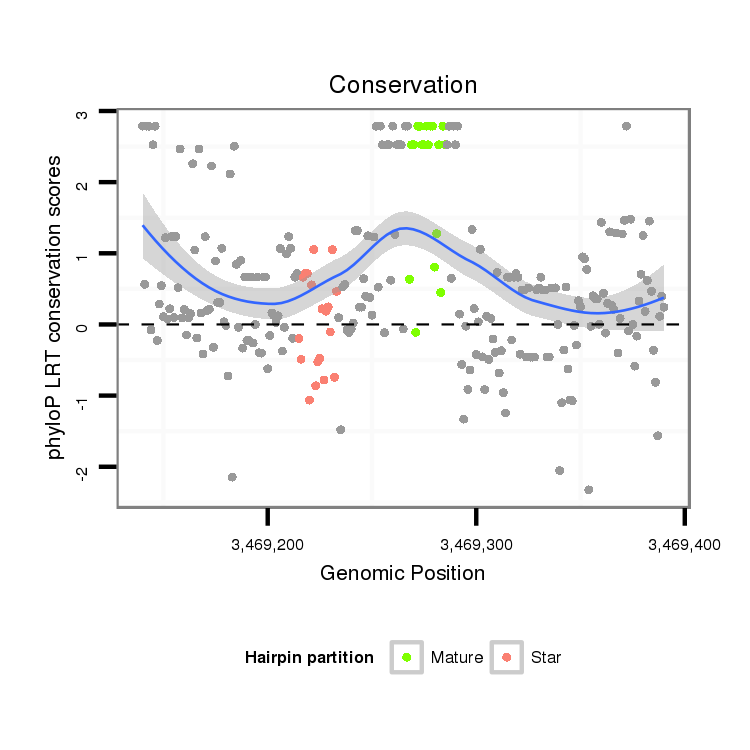
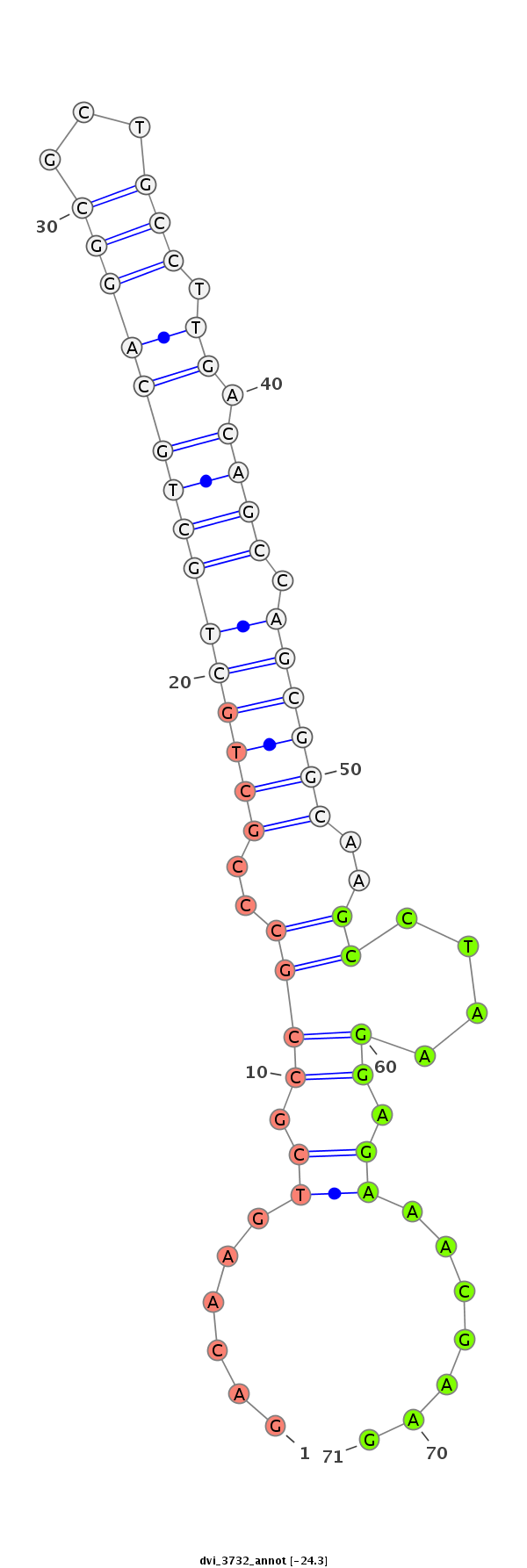
ID:dvi_3732 |
Coordinate:scaffold_12855:3469190-3469340 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.3 | -23.4 | -23.4 |
 |
 |
 |
CDS [Dvir\GJ10783-cds]; exon [dvir_GLEANR_10684:1]; intron [Dvir\GJ10783-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAAAGCAACGGAGCGCCGCTGCCGCTGCTGCAGCGACGTTCGCGTCAAGCGCCAACGTCCCTTGTCAACGCTGGAGACAAGTCGCCGCCCGCTGCTGCTGCAGGCGCTGCCTTGACAGCCAGCGGCAAGCCTAAGGAGAAACGAAGGTGAGTTATGATCATCCTAATTTGAAATCCCCCACACCTTTTTCTGCCAGATAGACTAATAAGAAATCTACGCTCCTGCCATGTGATCTCTGTTGGATGGGCTGT ***************************************************************************......((.((((..(((((((((((((((...))).)).))).)))))))..))....)).)).......********************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V053 head |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
SRR060670 9_testes_total |
M027 male body |
SRR060678 9x140_testes_total |
V116 male body |
GSM1528803 follicle cells |
SRR060660 Argentina_ovaries_total |
SRR060664 9_males_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060674 9x140_ovaries_total |
SRR060689 160x9_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060680 9xArg_testes_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................CCTAAAGAGTAACGGAGG........................................................................................................ | 18 | 3 | 20 | 2.95 | 59 | 0 | 0 | 18 | 21 | 3 | 2 | 1 | 1 | 0 | 0 | 3 | 0 | 5 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ................................................................................................................................GCCTAAGGAGAAACGAAG......................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AAAGCAACGGAGCGCCGCTGC..................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGGAGCACCGGTGCCGCTG................................................................................................................................................................................................................................ | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................ATACGCTCCTGCCATGTGATC................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GACAAGTCGCCGCCCGCTG............................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................GACAGCCAGCGGAAAGCCTAAGGAG................................................................................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGGAGCACCGGTGACGCTGCTG............................................................................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................ATGTGATCTCTGTTGGATGGG.... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TGCTGCAGCGACGTCGTCGTCAAGC......................................................................................................................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................GCTGCTGCAGGCGCTGCCT........................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GCCTTGACAGCCAGCGGAAAGCC........................................................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGATCGCTCTTGGTTGGGCT.. | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................GCGCTACACTGACAGCCAGCG............................................................................................................................... | 21 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GAGCGTCGCTGCCGCTGCTGC............................................................................................................................................................................................................................ | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGGAGCACCGGTGCCGCTGCCG............................................................................................................................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGGAGCACCGGTGACGCTGC............................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGGAGCACCGGTGCCGTTGC............................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AAAAGCTACGGGGAGCCGC........................................................................................................................................................................................................................................ | 19 | 3 | 13 | 0.23 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................CCTAAGGAGTAACGGAGGC....................................................................................................... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGAAGCACCGTTGCCGCTGCTG............................................................................................................................................................................................................................. | 22 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................CAGGTGAGTTTTGATGATCC........................................................................................ | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................ACAAGCCGCCGCCAGCTCCT........................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CAGCAGCGATGTTCGGGTC............................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................CCTAAGAAGTAACGGAGG........................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTTTCGTTGCCTCGCGGCGACGGCGACGACGTCGCTGCAAGCGCAGTTCGCGGTTGCAGGGAACAGTTGCGACCTCTGTTCAGCGGCGGGCGACGACGACGTCCGCGACGGAACTGTCGGTCGCCGTTCGGATTCCTCTTTGCTTCCACTCAATACTAGTAGGATTAAACTTTAGGGGGTGTGGAAAAAGACGGTCTATCTGATTATTCTTTAGATGCGAGGACGGTACACTAGAGACAACCTACCCGACA
*********************************************************************************************************......((.((((..(((((((((((((((...))).)).))).)))))))..))....)).)).......*************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060685 9xArg_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060679 140x9_testes_total |
GSM1528803 follicle cells |
SRR060662 9x160_0-2h_embryos_total |
V053 head |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................TGTTCAGCCGAGGGTGACGA........................................................................................................................................................... | 20 | 3 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................TTGCGACCTCTGTTCAGCGGC.................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGCGACCTCTGTTCAGCGGC.................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................TGTGGAAAAAGAAACTCTATC................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................CGGCTGAAGGGAACAGTT....................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................CCGACCTCTGTTCAACGACGG.................................................................................................................................................................. | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................TCCTTACAGCGGCGGGCGAC............................................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................ACGACCTGTGTTCAGCGACGG.................................................................................................................................................................. | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................CGGACGGTGCACTAGAGAA............. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:3469140-3469390 + | dvi_3732 | AAAAGCAA------CGGAGCGCC------G---CTGCCGCTGCTG------------CA---GCGACG---------TTCGCGTCAAGCGCCAACGTCCCTT---------------------------------------------GTCAACGCTGGAGACAA------------------------GTCGCCGCCC------------GCTGC------TGCTGCAGGCGCTGCCTTGACAGCCAGCGGCAAGCCTAAGGAGAAACGAAGGTGAGTTAT-G----AT-CATCCTAA----T-----------------------------TTGAAAT---------------------CC--CCCACACCTT---------TTTCTGCCAGATAGACTA-A------------TAAG-AAATC-TAC---------GCTC---------------CTGCC----------------------------------------------------------------------------ATGTGATCTCT-GTTGGATGGGCTG-T |
| droMoj3 | scaffold_6540:7770057-7770326 + | AAAAGCAA------CGGAGCGCC------G---CTGCCGCTGCTA------------CA---GCAACG---------TCG-------------------ACT---------------------------------------------GTCAGCGCTGGAGCCAAGTCACCGACCGCTGCTGCTGCTGGCGGTGT------TGTTGCTTCAGCAGC------AGCAGCAGGTATTGCCTTGACCGCAAGCGGCAAGCCTAAGGAGAAACGAAGGTGAGTTAT-G----GT-CACCCCAA----T---AG-----------------------T----TGGACTAATTTTGTCAACTCTCTTACCCCTCACT------AACA--------G--------CCAA-A------------TAAA-TAATC-TAA---------GCTC---------------CTGCC----------------------------------------------------------------------------ATGTGATCT-G-TCTGTCTGATGGG-C | |
| droGri2 | scaffold_14906:6150635-6150903 + | AAAAGCAA------CGGAGCGCC------G---CTGCCGCTGCAA------------CAGGAGCAACG---------TCGACGTCAAT-----------CT-----------CGTCC---------------------------------AAGGCTAGAACCAAGTCGACATCAGCTGCTGCTTCTGATTC------------------------------TGCTACAGGTGTGGGTTTGACTGCCAACGGCAAGCCTAAGGAGAAACGAAGGTGAGTTGATG----GT-CACCCTAA----T---CG------AT--------CTCAT-------------------------------GCTCCCCACTCCCTCCAACATTCCCCCTGCCGAATA-------GGT--------------AT-TC-AAAATTGACGACGCTC---------------CTGCC----------------------------------------------------------------------------ATGTGATCT-G--TCGGATGGGCTG-T | |
| droWil2 | scf2_1100000004943:5287856-5288017 + | AAAATCAG------CGCAATGGCGTGATCG---CTG------CTG------------------CATCTTCATCTCCATCGTCAT-------------------------------------------------------CAAGCT---------CT------------------------------GCCGCCAAGAAT------------TCAGGAGCATCCTCTGGAGGATCGGGACTAACTGCCAGCGGCAAGCCCAAGGAGAAACGCAGGTAAGTGTG-A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATCT---CTTAGCTGGATTT-T | |
| dp5 | 2:993203-993394 + | AAAAACAG------CGTAACGCC------G---CCGACCCCGCTG------------CT---GCTTCT---------------TCGAGCATCT-CTGCCACT---------------------------------------------GGCAGCGC------------------------------------GCT------AGCCAAGGCTTCTCC------AGGATCAGGATCGAACCTGACTGCCAGCGGCAAGCCCAAGGAGAAACGCAGGTGAGTCCC-C-----------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGCGTCCCGCC------------------------------C-------------AAT------CACCCACC--------------AATCGGATGT-C-TTTAAATCAGTTG-C | |
| droPer2 | scaffold_7:3779490-3779681 + | AAAAACAG------CGTAACGCC------G---CCGACCCCGCTG------------CT---GCTTCT---------------TCGAGCATCT-CTGCCACT---------------------------------------------GGCAGCGC------------------------------------GCT------AGCCAAGGCTTCTCC------AGGATCAGGATCGAACCTGACTGCCAGCGGCAAGCCCAAGGAGAAACGCAGGTGAGTCCC-C-----------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGCGTCCCGCC------------------------------C-------------AAT------CACCCACC--------------AATCGGATGT-C-TTTAAATCAGTTG-C | |
| droAna3 | scaffold_13340:20874368-20874608 + | AAAAACAAAAGCAGTCGG------CGATCG---CTCAATCCTCCT------------------------------------CTT----------------------------CGTCGACCACCCTTACTCCACCATCTT---CCTCT---------------------------------------GGATCCTCACC---------------CGT------CCTGGGAAAATCGAACCTCACGGCCAGCGGAAAACCCAAGGAGAAACGCAGGTGAGTCAT-A----GATC-----GGT------TAGGGTTAACTTTATATTCCTATAGG-C----------------------------------------------------------------------TGG--------------AG-TC--------------------------------CAC-------------------------------CC-------------CAT--ACCCGCCCCCC-----CGATATGCTCAATTGGATGT-T-TTTAGATCAGCGG-T | |
| droBip1 | scf7180000396413:1531041-1531290 - | AAAAGCAAAAGCAGTCGG------CAATAG---CTGCATCCACCT---C------CT------CGTC---CTCCT------CAT----------------------------CGTCGACCACCA---CCTCGCCATCTTCCT---CC---------------------------------------GGATCCGGATCA------------CCCGT------CCTGGTCAAATCGAACCTCACGGCCAGCGGCAAGCCCAAGGAGAAACGCAGGTGAGTCAT-A----GATCTA---GA----TGTTAG------AT--------CTATA---------------------------------------------------------------------G-ACCTGG--------------AG-TC-----------------------------------------------------CCTATACCCCCACCC-------------TAT-ACACCCTAC-----------CATGCTCTATTGGATCT-TTTTTAGATCAGAGG-T | |
| droKik1 | scf7180000302261:327876-328084 + | AGAAGCAA------CTGG------CGGCTG---CTGCATCCACCT------------CA---ACATCT----------------------------------------------TCCTCGGCCT---C---ATCATCGGCCTCTCCT---------------------------------------GTTTCCTCCGAT------------TCCTC------GGCCTCCAAATCCAATCTCACGGCCAGCGGAAAGCCAAAGGAGAAACGAAGGTGAGTGGA-A----TG-------CA----A-----------------------------C----------------------------------------------------------------------C---CAAATTACACTGT------------------------------------------------------------------GACCATA-------------TCC-ATTGCGATCC------------------CTCGAATAT-ATTTTAGATCAATTGCT | |
| droFic1 | scf7180000454073:3457463-3457692 - | AGAAGCAAACCGAGTCCG------TGGCTT---CTAACTCCTCC------------------------------T------CTT----------------------------CGATTTCCTCCT---CCCCACCGTCTTCGTCCTCT---------------------------------------GGCTCCCCATCT------------TCTGC------CGCCCCCAAATCCAATCTCACGGCCAGCGGAAAACCAAAGGAGAAGCGAAGGTGAGTCAC-A----CT-------AC----------------------------------C------------------------------------------------------------------------------------AATAA-TC-CGA---------TCCC---------------------------------------------TTAAACGATAACCTAATCCAATA-ATACCATC--------TGAAGCGCTCAAATGGATCC-A-AT----GGGCCTG-C | |
| droEle1 | scf7180000491280:2165593-2165845 + | AGAAGCAG---CAATCGG------TA---------------------------------------------TCCT------CAT----------------------------CGTATTCCTCCT---CCCCACCATCTTCGTCTTCTAGT------------------------------------GGTTCCCT------CTCACCACCCTCTGC------CGCCCCAAAATCCAATCTCACGGCCAGCGGTAAACCAAAGGAGAAACGAAGGTGAGTCAG-A----CA-CAAACAAA----T-----------------------------CCTACAT----------------------------------------------------------AGCT--TGC-----------AATAA-TT-CGA---------TCTC---------------------------------------------CTGACCAATACACCCATCCAACA-ATCCAATCCG---ATCCGTAGTACTCCATTGGATCT-G-CTTAGATCACTTG-T | |
| droRho1 | scf7180000779336:86712-86958 - | AGAAGCAA------TCGG------TGGCTGCCACCGACTCCTCCT------------------------------------CTT----------------------------CTTATTCCACCT---CTTCACCCTCTTCGTCTTCT---------------------------------------GGCTCTCT------TTCATCATCCCCTGC------CGCCCCAAAATCCAATCTCACGGCTAGCGGTAAACCAAAGGAGAAACGAAGGTGAGTCGG-A----CA-------GATAAAC----------------------------------AT----------------------------------------------------------GCCA-T------------CAAA-AAATC-AGA---------TCCC---------------------------------------------CTGACCAATACACCCATCCAATA-ATCCGATC--------CGTAGTGCTCCCTTGGATCT-G-CTCAGATCAC----T | |
| droBia1 | scf7180000302113:1508085-1508338 - | AGAAGCAG------TCGG------TGGCCA---CCAACTCCGCCTCCTC---CTCCG------CCTC---ATCCT------CCT----------------------------CGTACTCCTCCT---CCCCACCATCTTCGTCTGCG---------------------------------------GGATCCCCATCT------------TCGGC------GGCCCCAAAGTCCAATCTCACGGCCAGCGGCAAACCGAAGGAGAAACGAAGGTAAGTCAT-AGTGATC-------GATAG------------------------------A----TGA----------------------------------------------------------GCTA-T------------CTGATTA-TCCGCA---------TACCCTGACCCAGAGTATCT--------------------------------------------------------CGATCCCCCAT-----CCGAATCCATCGGATCT-G-CCTAGATCAG----T | |
| droTak1 | scf7180000415765:297992-298261 + | AGAAGCAA------TCGG------TGGCCA---CCAATTCCTCCTCCTC------CT------CC---------T------CCT----------------------------CGTTTTCTTCCT---CCCCACCATCTTCGTCTTCT---------------------------------------GGCTCCCCATCTTCTTCCTCATCTTCGGC------AGCCCCAAAATCCAATCTCACGGCCAGCGGCAAACCAAAGGAGAAACGAAGGTGAGTCAT-GTGGATC-------GAAAG------------------------------A----TAT----------------------------------------------------------AGTA-C------------TGGATAA-TCCTGA---------TACCCAGACCCACAGTATCCC-------------------------------------------ATTCAATT-ATGCGATCCCCCGAT-TGTA--ATTCCATCGGCTCT-G-CTTAAATCGC----- | |
| droEug1 | scf7180000409555:366545-366784 + | AAAACCAATCGCAGTCGG------TGGCCA---CGAGCTCCTCCACCTC------CT------CC---------C------CCT----------------------------CGTTTTCAACCT---CGCCATCATCTTCGTCTTCT---------------------------------------GGCTCTCCATCTACCTCCTCATCCTGCGC------GGCCCCAAAATCCAATCTCACGGCCAGCGGCAAACCGAAGGAGAAGCGAAGGTGAGTCAC-T----TG-------AA----C-----------------------------C------------------------------------------------------------------------------------------------------------------------------GCTGTACTGTCCACCAATCCGAATATC-TTGATC---------------G-AATTCC----------------------ATCCGATTT-G-CTTAGATCAGATG-T | |
| dm3 | chr3R:25884690-25884924 + | AGAAGCAGTCCCAAGCGG------TGGTCA---CCAACACCTCCGCCTCCTCCTCCT------CCTG---TTCCT------CTT----------------------------CGTTCTCGTCCT---CCCCACCATCTTCGTCCGTT---------------------------------------GGCTCCCCATCT------------CCTGG------CGCCCCAAAAACCAATCTCACGGCCAGCGGCAAACCAAAGGAGAAAAGAAGGTGAGTCAC-A----CG-------AA----T-----------------------------C----------------------------------------------------------------------TGC--------------AA-TC-----------------------------------------------------CGGATACCGCCGATC-------------TAC-AATCCCATCCGTT--------------GGTTGGATCT-G-CCTAGATCGC----- | |
| droSim2 | 3r:25230890-25231124 + | AGAAGCAGTCCCAAGCGG------TGGCCA---CCAACTCCTCCGCCTCCTCCTCCT------CCTG---CTCCT------CCT----------------------------CGTTCTCGGCCT---CCCCACCATCTTCGTCCGTT---------------------------------------GGCTCCCCATCT------------CCTGG------CGCCCCAAAAACCAATCTCACGGCCAGCGGCAAACCAAAGGAGAAACGAAGGTGAGTCCC-A----CG-------AG----T-----------------------------C----------------------------------------------------------------------CGC--------------AA-TC-----------------------------------------------------CGGATACCGCCGATC-------------TAC-AATCCCATCCCCT--------------GGTTGGATCT-G-CCTAGATCGC----- | |
| droSec2 | scaffold_4:4730666-4730906 + | AGAAGCAGTCCCAAGCGG------TGGCCA---CCAACTCCTCCGCCTC------CT------CCTC---CTCCT------CCT----------------CCTGCTCCTCTATGTTCACGGCCT---CCCCACCATCTTCGTCCGTT---------------------------------------GGCTCCCCATCT------------CCTGG------CGCCCCAAAAACCAATCTCACGGCCAGCGGCAAACCAAAGGAGAAACGAAGGTGAGTCAC-A----CG-------AA----T-----------------------------C----------------------------------------------------------------------TGC--------------AA-TC-----------------------------------------------------CGGAAACCGCCGATA-------------TGC-AATCCCATCCTCT--------------TGTTGGATCT-G-CCTAGATCGC----- | |
| droYak3 | 3R:26807892-26808131 + | AGAAGCAGTCCCAATCGG------TGGCCA---CCAACTCCTCCTCCTC------CT------CCTC---ATCCT------CTT----------------------------CGTTCTCGGCCT---CCCCGCCATCTTCGGCCGTT---------------------------------------GGCTCCCCAGCT------------CCTGG------CGCCCAAAAATCCAATCTCACCGCCAGCGGCAAACCAAAGGAGAAACGAAGGTGAGTCAC-A----TG-------GA----C-----------------------------T-----A----------------------------------------------------------TGTA-C------------ATGGTAA-CC-----------------------------------------------------CGGATGCC-CTGATC-------------TAT-AATCCCATCCCCA--------------AATTCGATCT-G-CCTAGATCGCTTG-T | |
| droEre2 | scaffold_4820:2114817-2115052 - | AGAAGCAGTCCCAATCG---------GCCA---CCAACTCCTCATCCGC------CT------CC---------T------CCT----------------------------CGTTCTCGGCCT---CCCCCACATCTTCGTCCGCT---------------------------------------GGCTCTCCATCT------------CCTGG------CGCCCCGAAATCCAATCTCACGGCCAGCGGCAAACCAAAGGAGAAACGAAGGTGAGTCAC-A----TG-------GA----T-----------------------------C----------------------------------------------------------------------TGCCTATACTACATGGTAA-TC-----------------------------------------------------CGGATGCC-CTCATC-------------TAT-AATCCCAGCCCTT--------------GATTGGATCT-G-CCTAGATCGC-GC-T |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 09:03 PM