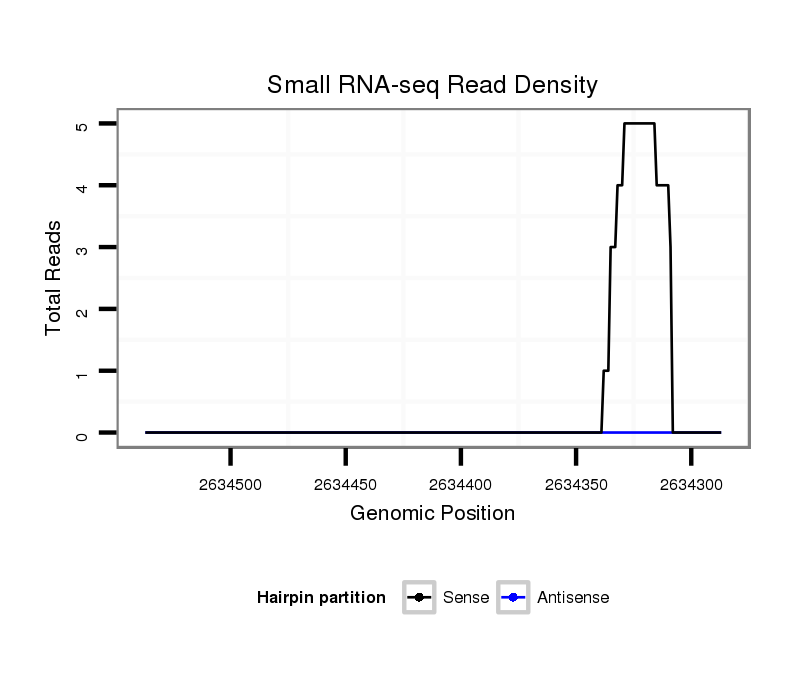
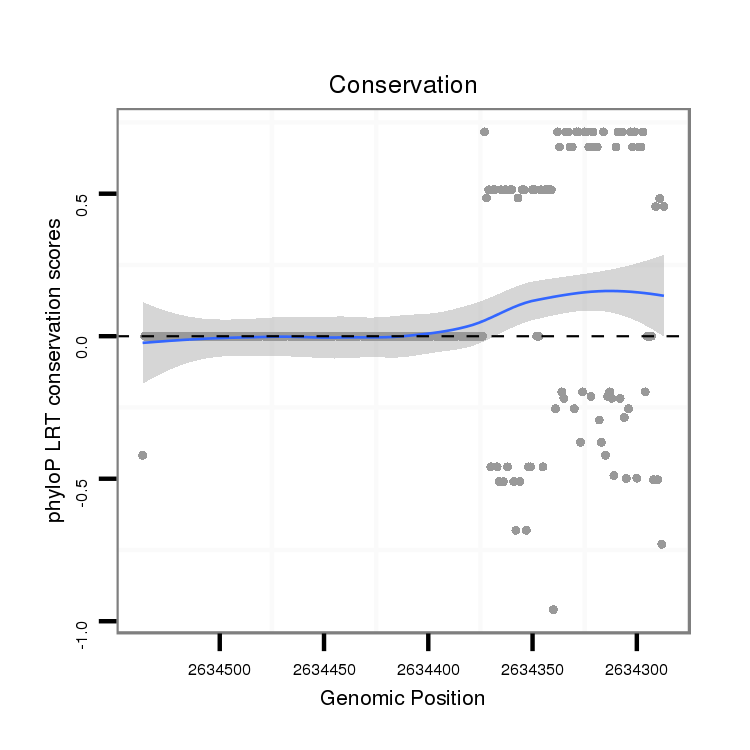
ID:dvi_3597 |
Coordinate:scaffold_12855:2634337-2634487 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ10448-cds]; exon [dvir_GLEANR_10373:4]; intron [Dvir\GJ10448-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAATTATATTAAAAACATAAGAATATTATGAATACGTGTTTCAGCGGAAATTCCACTCGCAGCAGTGGTAAATTAGCGGAAAACTTTTGTATAAAATTTGTTTGTTTCCCTTTCGATCAGTTACGAAAACATTTTTGAGAACATTGATTTTATAAGTTAATTTTTCAGAATATAATAAATGTATTTGTTTATGTTTTTTAGCATTCCTATCGTTGTACCCGTAACGCCTGTAATTGATCCTGAACCCGTAG **************************************************..(((((......)))))......((.((((.((...................))))))))......((...(((((.(((((((((((((.((((((((...))))))))))))))))........))))).)))))...))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060682 9x140_0-2h_embryos_total |
M027 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
GSM1528803 follicle cells |
M047 female body |
M061 embryo |
SRR060666 160_males_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060660 Argentina_ovaries_total |
V116 male body |
SRR060665 9_females_carcasses_total |
SRR060656 9x160_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................GAAATAGATCCTGAAACCGT.. | 20 | 3 | 8 | 17.88 | 143 | 29 | 22 | 2 | 22 | 11 | 16 | 8 | 7 | 0 | 0 | 0 | 3 | 3 | 5 | 3 | 2 | 2 | 1 | 3 | 1 | 0 | 1 | 1 | 1 |
| ....................................................................................................................................................................................................................................TGAAATAGATCCTGAAACCGT.. | 21 | 3 | 5 | 3.20 | 16 | 2 | 4 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................AATAGATCCTGAAACCGT.. | 18 | 2 | 8 | 3.00 | 24 | 4 | 6 | 1 | 4 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................AGCATTCCTATCGTTGTACCCGTAACGCC....................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CCTATCGTTGTACCCGTAACGCCT...................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AAGTTAATTTTTCGGAAT................................................................................ | 18 | 1 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ATTCCTATCGTTGTACCCGT............................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................ATCGTTGTACCCGTAACGCCT...................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ATTCCTATCGTTGTACCCGTAACGCCT...................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GAAATAGATCCTGAACCCGT.. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................AAATTAGCGGATGACTTGTG.................................................................................................................................................................. | 20 | 3 | 10 | 0.30 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GATTCTTCAGAATATAATA.......................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................ATTAGCGGATGACTTGTG.................................................................................................................................................................. | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GAAAACCTATCGTTGTACC................................ | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................AATTAGCGGATGACTTGTG.................................................................................................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTTAATATAATTTTTGTATTCTTATAATACTTATGCACAAAGTCGCCTTTAAGGTGAGCGTCGTCACCATTTAATCGCCTTTTGAAAACATATTTTAAACAAACAAAGGGAAAGCTAGTCAATGCTTTTGTAAAAACTCTTGTAACTAAAATATTCAATTAAAAAGTCTTATATTATTTACATAAACAAATACAAAAAATCGTAAGGATAGCAACATGGGCATTGCGGACATTAACTAGGACTTGGGCATC
**************************************************..(((((......)))))......((.((((.((...................))))))))......((...(((((.(((((((((((((.((((((((...))))))))))))))))........))))).)))))...))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060687 9_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060676 9xArg_ovaries_total |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................................AAAAAAGGGAAAGCAAGT.................................................................................................................................... | 18 | 2 | 20 | 0.10 | 2 | 0 | 1 | 1 |
| .......................................................................................................................................................................................................TCGTAAGGAGAGCATGATG................................. | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:2634287-2634537 - | dvi_3597 | AAATTATATTAAAAACATAAGAATATTATGAATACGTGTTTCAGCGGAAATTCCACTCGCAGCAGTGGTAAATTAGCGGAAAACTTTTGTATAAAATTTGTTTGTTTCCCTTTCGATCAGTTACGAAAACATTTTTGAGAACATTGATTTTATAAGTTAATTTTTCAGAATATAATAAATGTATTTGTTTATGTTTTTTAGCATTCCTATCGTTG---TACCCGTAACGCCTGTAATTGATCCTGAACCCGTAG |
| droMoj3 | scaffold_6540:11834382-11834434 - | T-------------------------------------------------------------------------------------------------------------------------------------------------------------------T--------------------------------ACAGCATTCCCATTGTTG---TACCCATTACACCTATGCCTGAGCCTG---TCATCG | |
| droGri2 | scaffold_14624:2311433-2311512 + | T-------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAAAACTTTACAATGGAATGCATT--TATTTTCTAGTTTTCCTATTATTGAAGCACCAATTGTGGCTGTGATTGATCCTA--------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 07:54 PM