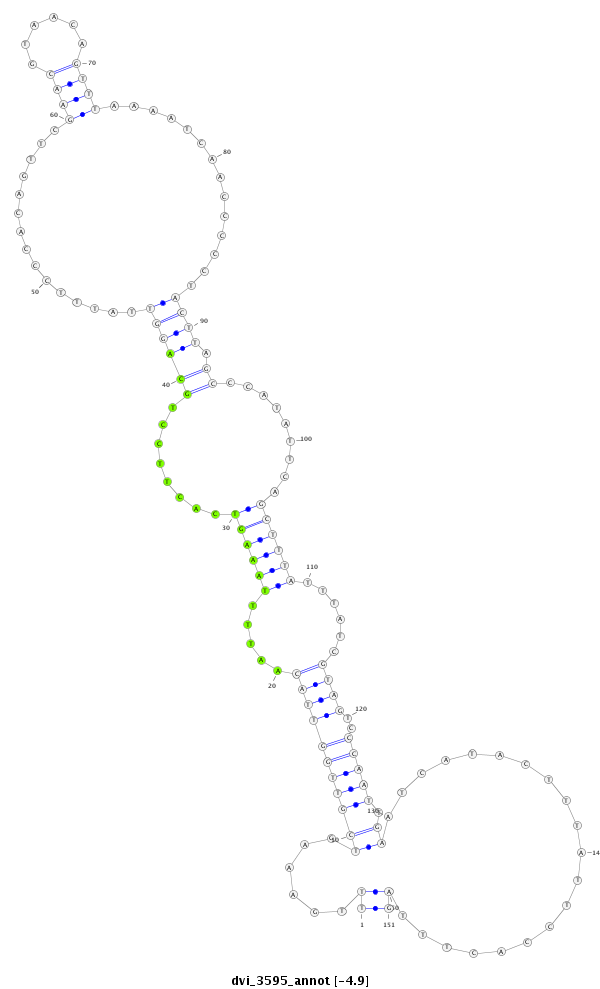
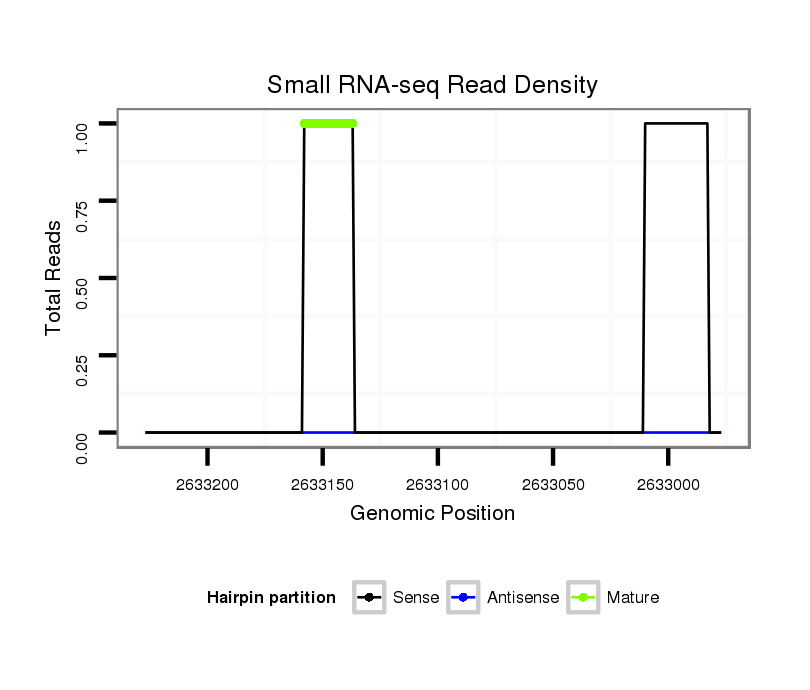
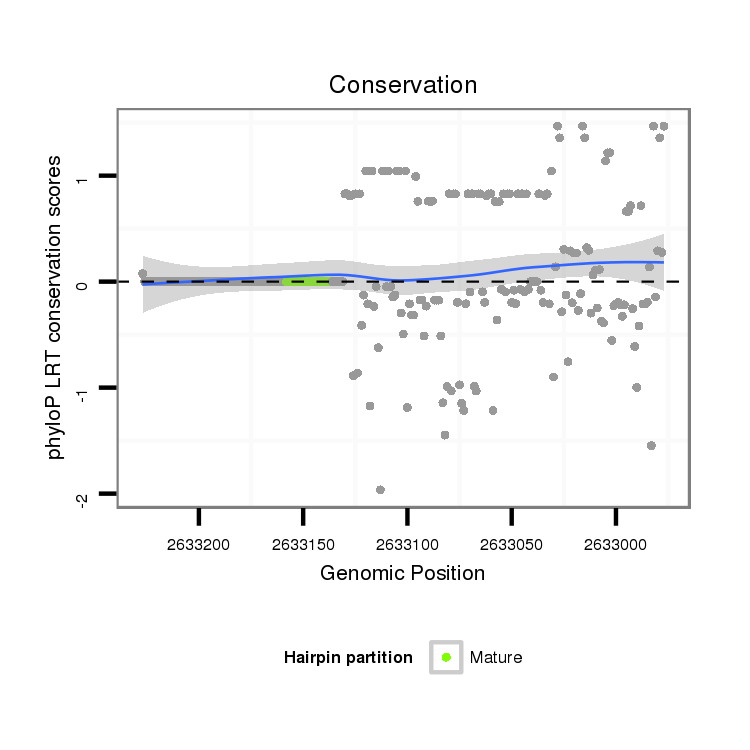
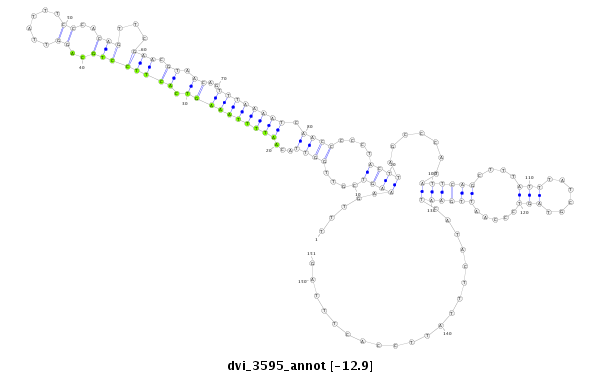
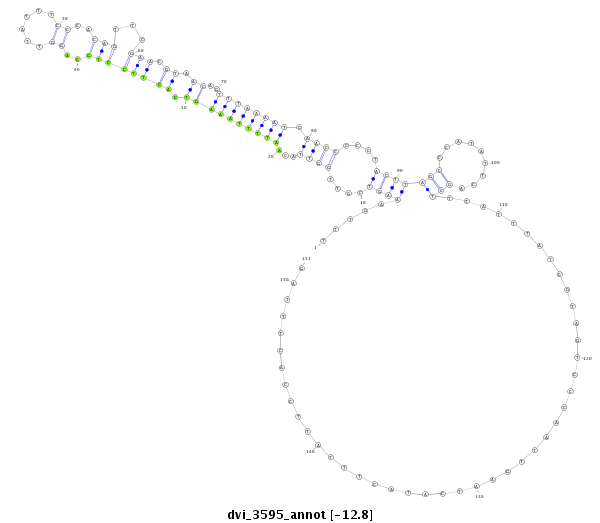
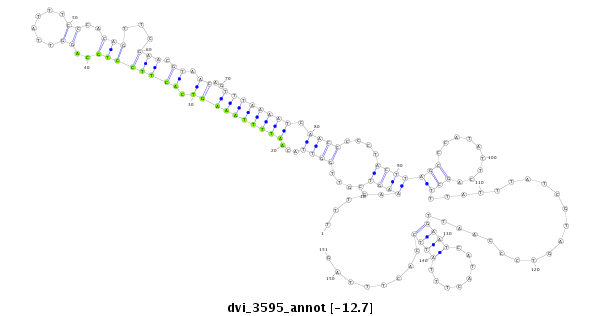
ID:dvi_3595 |
Coordinate:scaffold_12855:2633027-2633177 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.9 | -12.8 | -12.7 | -12.7 |
 |
 |
 |
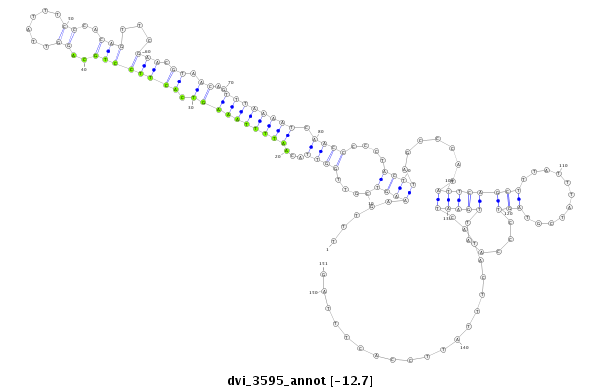 |
CDS [Dvir\GJ10448-cds]; exon [dvir_GLEANR_10373:7]; intron [Dvir\GJ10448-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGATTTCTGTCGAATTTCCTTTCAACAATTCACTTTACCTAAAATTACAGTTTGAAAGTCGTTGGTTACAATTTTAAAGTCACTTCCTGCAGGTTATTTCCCACAGTTCGAACGTAACAGTTTAAAATCAACCCCCTACTTAGCCCATATTCAGCTTTATTTATCGTAGTCCCAATTGAATCATACTTTATTCCACTTTAGCCTCCACCACACGTACAGTATGATATACAGCCCATGCTTTACTTTGCCGA **************************************************((......(((((((((((.....((((((........((((((...............((((......))))..............)))).)).........))))))......))))..))))).))....................))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M061 embryo |
SRR060671 9x160_males_carcasses_total |
GSM1528803 follicle cells |
M027 male body |
V053 head |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................AGTATGATATACAGCCCATGCTTTACTT...... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................AATTTTAAAGTCACTTCCTGCA................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CGTAACGGTTTAAATTCA......................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..TTTGCTGTCGAATTTCCTTT..................................................................................................................................................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TGAAAGGCGTTGGTTGCAA.................................................................................................................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................CAGCCCATCCTTCACGTTGC... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................CACATTCCCTAAAGTTACAG......................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................AGCTTGAAAGGCGTTGGT......................................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................ATGAAAGTCGTTGGTT........................................................................................................................................................................................ | 16 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................AACAGCGTAAAAGCAACCCC.................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................CAAAATTGCAGTTTGAAA.................................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TCTAAAGACAGCTTAAAGGAAAGTTGTTAAGTGAAATGGATTTTAATGTCAAACTTTCAGCAACCAATGTTAAAATTTCAGTGAAGGACGTCCAATAAAGGGTGTCAAGCTTGCATTGTCAAATTTTAGTTGGGGGATGAATCGGGTATAAGTCGAAATAAATAGCATCAGGGTTAACTTAGTATGAAATAAGGTGAAATCGGAGGTGGTGTGCATGTCATACTATATGTCGGGTACGAAATGAAACGGCT
**************************************************((......(((((((((((.....((((((........((((((...............((((......))))..............)))).)).........))))))......))))..))))).))....................))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060687 9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060668 160x9_males_carcasses_total |
M047 female body |
SRR060663 160_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060664 9_males_carcasses_total |
M061 embryo |
V053 head |
SRR060679 140x9_testes_total |
SRR060672 9x160_females_carcasses_total |
M027 male body |
SRR060659 Argentina_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................TAGGTGAAATGGATATCAAT............................................................................................................................................................................................................ | 20 | 3 | 6 | 3.67 | 22 | 2 | 0 | 4 | 6 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............AGGGAAAGTTGTTAAGTGCA........................................................................................................................................................................................................................ | 20 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AAGGGAAAGTTGTTAAGTG.......................................................................................................................................................................................................................... | 19 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GGGAAAGTTGTTAAGTGCA........................................................................................................................................................................................................................ | 19 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GGAAAGTTGTTAAGTGCA........................................................................................................................................................................................................................ | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GGAAAGTTGTTAAGTGTA........................................................................................................................................................................................................................ | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................GAGAAGGACGTCCATTAA......................................................................................................................................................... | 18 | 2 | 6 | 0.50 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TAGGTGAAATGGATATCAA............................................................................................................................................................................................................. | 19 | 3 | 20 | 0.50 | 10 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| .............GAAGGGAAAGTTGTTAAGTG.......................................................................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AGGGAAAGTTGTTAAGTGCAT....................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AAAAGGAAGTTGTTAAGTG.......................................................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CAGTGAAGAACGTCCAAG........................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................ACTGTATGTGGGGTACGGAAT......... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TAAGTTGTTAAGTGTAATGCAT.................................................................................................................................................................................................................. | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AGGGAAAGTTGTTAAGTGC......................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GGAAAGTTGTTAAGTGCAT....................................................................................................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TTAGGTGAAATGGATATCAA............................................................................................................................................................................................................. | 20 | 3 | 13 | 0.15 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GAAGGGAAAGTTGTTAAG............................................................................................................................................................................................................................ | 18 | 2 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AAGGGAAAGTTGTAAAGTGCA........................................................................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CAGAGAAGGACGTCCATGA.......................................................................................................................................................... | 19 | 3 | 17 | 0.12 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AAGATGTTAAGTGTAATG..................................................................................................................................................................................................................... | 18 | 2 | 17 | 0.12 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................ATACAAAATGTCGAGTACGAAA.......... | 22 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................AGGTGAAATGGATATCAAT............................................................................................................................................................................................................ | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TTATTAGGTGAAATGGATAT................................................................................................................................................................................................................ | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................GGAAAGTTGTTAAGTGCATG...................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................CGGGGTTAACCTAGTATGG................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............AAAGGAAGTTGTTAAGTG.......................................................................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............AAAAGGAAGTTGTTAAGTGG......................................................................................................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TCGAACAAACTAGCATCAG................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AGGGAAAGTTGTAAAGTG.......................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................AGGGAAGGACGTCCATGAA......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:2632977-2633227 - | dvi_3595 | AGATTTCTGTCGAATTTCCTTTCAACAATTCACTTTACCTAAAATTACAGTTTGAAAGTCGTTGGTTACAATTTTAAAGTCACTTCCTGCAGGTTATTTCCCACAGTTCGAACGTAACAGT-----TTAAAATCAACCCCCTACTTAGCCCA----------TATTCAGCTTTATTTATCGTAGTCCCAATTGAATCA---TACTTTATTCCACTTTAGCCTCCACCACACGTACAGTATGATATACAGCCCATGCTTTACTTTGCCGA |
| droMoj3 | scaffold_6540:11833290-11833378 - | A--------------------------------------------------------------------------------------------------------TCTTAATTAAAATAAAATCTCCTACTACTCCC----------------------------------------------------------------------------TTGTAGCCTGAGCCTTACATACCGTTGGATTTACATTCCACCAATTACAATAACGA | |
| droGri2 | scaffold_14624:2312353-2312405 + | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATACCACCTCACGTCTATTTTGATGTGTTGCCCATGTATCGTTTTGCCGA | |
| droBip1 | scf7180000396728:712456-712483 + | T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGGCCACGTTCTACGAGT---------------------------ATACCGA | |
| droYak3 | 2L:64081-64230 + | T------------------------------------------------------------------------------------------------TTCCCATATTTTGATCCGAACAGTGCCTTTTACCACTAACCTTGAACTCGAGAGATGAAAAACTGTTTTAGTTGTAATATAAAGTAA---TATTTTGCTTATAAAAT----TCACATTCTAGGCCTCATTTTACGAATATGCAGATGC----------------TTTGCCAA | |
| droEre2 | scaffold_4929:102138-102265 + | T------------------------------------------------------------------------------------------------TTCCTACATTTTAATCCCAACAGTGCCTTTTACCATTAAC------------CTG----------TATTAAGAGTAACTTAAAGTATTACTATTTTGCTTA---AAT----TCACATTCTAGGCCTCATTTTACGAATATGCAGATGA----------------TGTGCCAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 07:54 PM