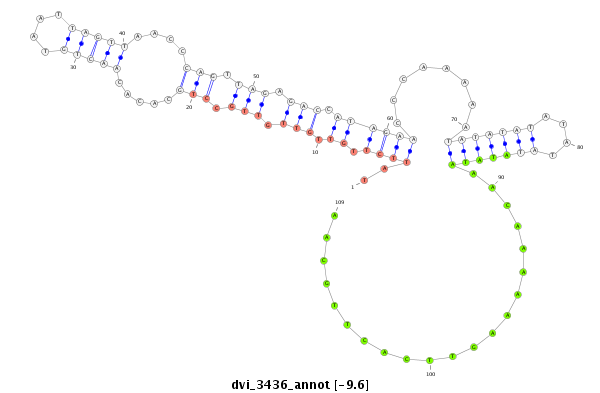
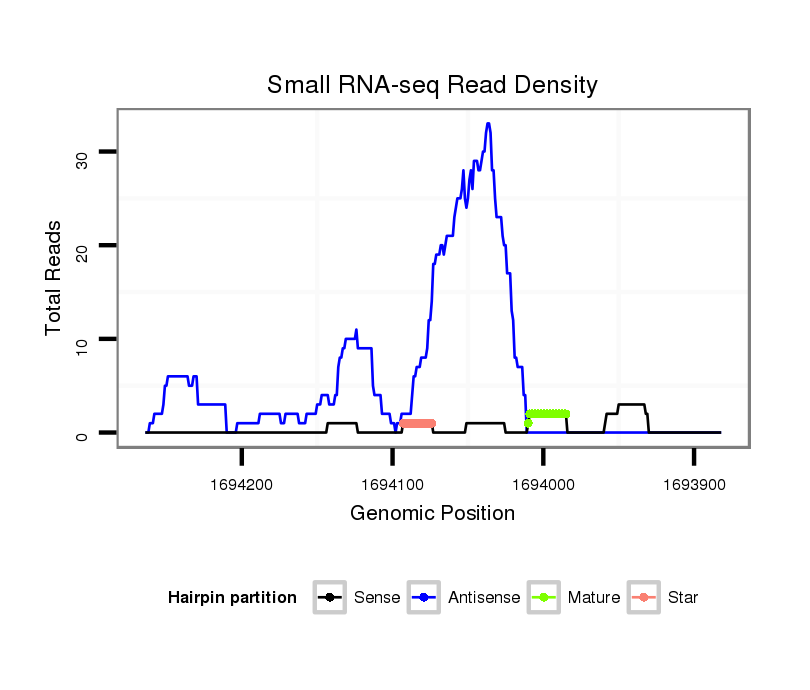
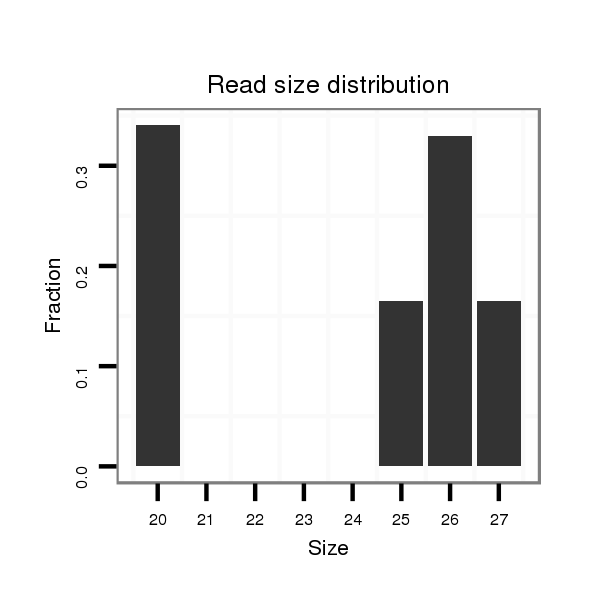
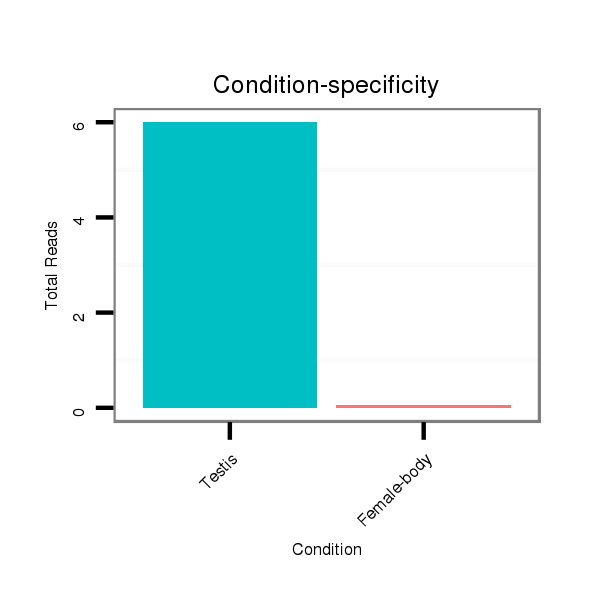
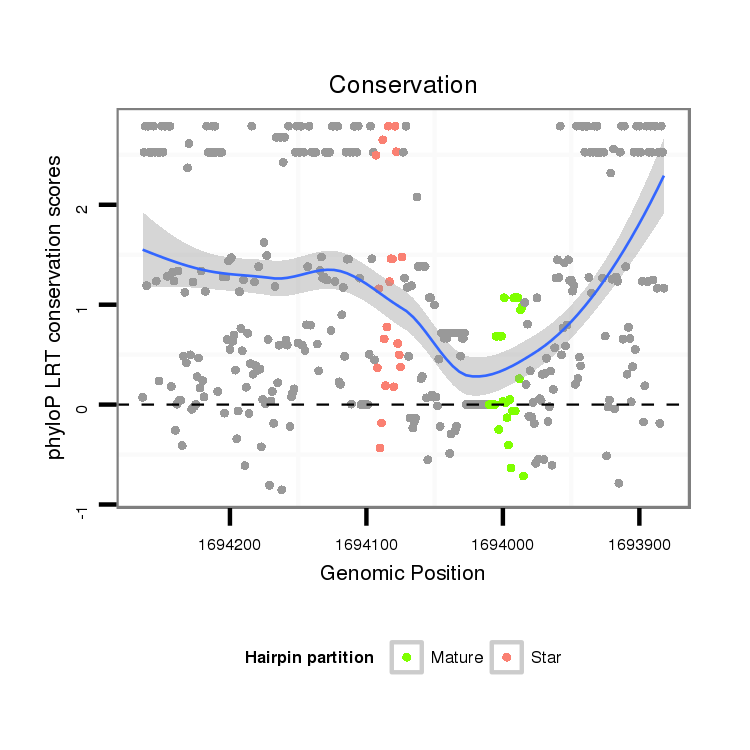
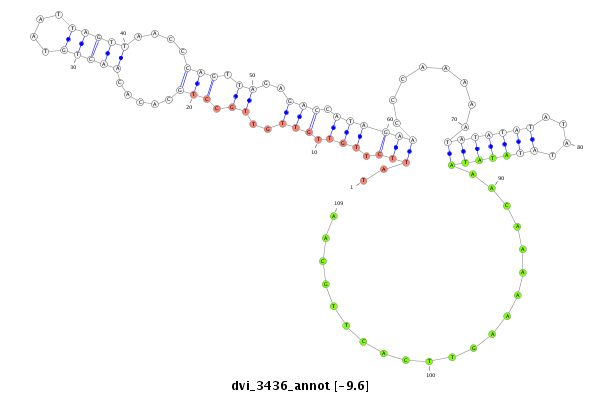
ID:dvi_3436 |
Coordinate:scaffold_12855:1693932-1694214 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.6 | -9.6 | -9.6 |
 |
 |
 |
CDS [Dvir\GJ10495-cds]; exon [dvir_GLEANR_10416:8]; CDS [Dvir\GJ10495-cds]; exon [dvir_GLEANR_10416:7]; intron [Dvir\GJ10495-in]
| Name | Class | Family | Strand |
| (AGTTG)n | Simple_repeat | Simple_repeat | + |
| AT_rich | Low_complexity | Low_complexity | + |
| (CAT)n | Simple_repeat | Simple_repeat | + |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CGTCTGGGAGCGGCACAAGAAACGTCTCAGCAGCAGCGCTGACAACCAGGGTGAGGCGGGTGGACACAATGATGGTGATGATGTTGATCATGAAGTTGATGATGATGATCATGATGTGCAAATTGGGGCACAGACACGCTACTAACTGGCAACTGTAGTCCTAGTTGTTGTTATTCTTGTTGTTGTTGCCTGCACACAACTGTAATTAGTTAACCCAGTTAGAGACCATAGAACCCAAAAATATATATATATATATATAAACAAAAAAGTTCACTTGCAATTCAACTGAACCCAACTGAACTCAACTGAACTTCCTCCAAACATCTTCCGCAGAGGCCCGGAGCTATGCTGACTTTAGCCCCAGTCCCATGCACGATGCTTCC ***************************************************************************************************************************************************************************..((((.((.(((.(((.(((.....(((((....)))))....))).))).))).))))))........(((((((....))))))).....................******************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060680 9xArg_testes_total |
M047 female body |
SRR060665 9_females_carcasses_total |
SRR1106729 mixed whole adult body |
GSM1528803 follicle cells |
SRR060686 Argx9_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................ATATAAACAAAAAAGTTCACTTGCAA....................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CCAGGGTGAGGCGGGAGA................................................................................................................................................................................................................................................................................................................................ | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TATTCTTGTTGTTGTTGCCT................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................ATTGGGGCACAGACACGCTA.................................................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................TCAACTGAACCCAACTGAACTTCCTCCA................................................................ | 28 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................CTCCAAACATCTTCCGCAGA................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CTGAACTTCCTCCAAACATCTTCCGCA................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................TGAACTTCCTCCAAACATCTTCCGCAGA................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CCCAGTTAGAGACCATAGAACCCAAA................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................TATAAACAAAAAAGTTCACTTGCAA....................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTGAGGCCGGTAGACACAC.......................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................ATGGTGATGATGTTGATCTCG................................................................................................................................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................ATTCTGACTGAAGCCCCAGT.................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................CCCAGGGTGAGGCGGGAGA................................................................................................................................................................................................................................................................................................................................ | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................CAGAGGCCAGCACCTATGCT................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................TTGTTATTCTTGTTGTTGTT.................................................................................................................................................................................................... | 20 | 0 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................CACCCAGGGTTAGTCGGG................................................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................AGTCAGAGAGCATAGAACC.................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCAGACCCTCGCCGTGTTCTTTGCAGAGTCGTCGTCGCGACTGTTGGTCCCACTCCGCCCACCTGTGTTACTACCACTACTACAACTAGTACTTCAACTACTACTACTAGTACTACACGTTTAACCCCGTGTCTGTGCGATGATTGACCGTTGACATCAGGATCAACAACAATAAGAACAACAACAACGGACGTGTGTTGACATTAATCAATTGGGTCAATCTCTGGTATCTTGGGTTTTTATATATATATATATATATTTGTTTTTTCAAGTGAACGTTAAGTTGACTTGGGTTGACTTGAGTTGACTTGAAGGAGGTTTGTAGAAGGCGTCTCCGGGCCTCGATACGACTGAAATCGGGGTCAGGGTACGTGCTACGAAGG
*******************************************************************************************************..((((.((.(((.(((.(((.....(((((....)))))....))).))).))).))))))........(((((((....))))))).....................*************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060680 9xArg_testes_total |
SRR060659 Argentina_testes_total |
V053 head |
M061 embryo |
V116 male body |
M047 female body |
SRR060660 Argentina_ovaries_total |
M027 male body |
M028 head |
SRR060668 160x9_males_carcasses_total |
GSM1528803 follicle cells |
SRR060665 9_females_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060670 9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................CGTGTGTTGACATTAATCAATTGGGT...................................................................................................................................................................... | 26 | 0 | 1 | 4.00 | 4 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................GTGTCTGTGCGATGATTGACCGT........................................................................................................................................................................................................................................ | 23 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................GTATCTTGGGTTTTTATATATATATAT.................................................................................................................................. | 27 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TAATCAATTGGGTCAATCTCTGGTAT......................................................................................................................................................... | 26 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................AATCAATTGGGTCAATCTCTGGTATCTT...................................................................................................................................................... | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GTGTTCTTTGCAGAGTCGTCGT............................................................................................................................................................................................................................................................................................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................ACGTGTGTTGACATTAATCAATTGGGT...................................................................................................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CAATCTCTGGTATCTTGGGTTTTTAT............................................................................................................................................ | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CGTCGCGACTGTTGGTCCCACT......................................................................................................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GTCAATCTCTGGTATCTTGGGTTTT............................................................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CTGGTATCTTGGGTTTTTATATATATC..................................................................................................................................... | 27 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................AATCTCTGGTATCTTGGGTTTTTATAT.......................................................................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GGACGTGTGTTGACATTAATCAAT........................................................................................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................TTGGGTCAATCTCTGGTATCTTGGGT.................................................................................................................................................. | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACAACAACAACGGACGTGTGTTGACAT................................................................................................................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................TATCTTGGGTTTTTATATATATATAT.................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................AATCAATTGGGTCAATCTCTGGTATCT....................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CGTTTAACCCCGTGTCTGTGCGAT.................................................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ATCAATTGGGTCAATCTCTGGTATCT....................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CTACTACAACTAGTACTTCAACTACT......................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................CTCTGGTATCTTGGGTTTTTATAT.......................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................ACATTAATCAATTGGGTCAATCTCT.............................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CTGCGCGATGATTGACCGTTGACAT.................................................................................................................................................................................................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............GTTCTTTGCAGAGTCGTCGT............................................................................................................................................................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................ACAACGGACGTGTGTTGACAT................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................GCATCGTTGACATCAGGATCAA......................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................GACATTAATCAATTGGGTCAATCTCTG............................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................CGCGACTGTTGGTCCCACT......................................................................................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TGGTATCTTGGGTTTTTATATATATAT.................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................CAACAACAACGGACGTGTGTTGACATT.................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................TGTCTGTGCGATGATTGACCGTT....................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CAATCTCTGGTATCTTGGGTTTTTATA........................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CGGACGTGTGTTGACATTAATCAAT........................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................AACAATAAGAACAACAACAACGGACGT............................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CAATCTCTGGTATCTTGGGTTTTTATAT.......................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................AACAACGGACGTGTATTGACAT................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................TCGGGGACAGGGTGCGCGCT....... | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................ATTGGGTCAATCTCTGGTATCTTGGGTT................................................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TAATCAATTGGGTCAATCTCTGGTA.......................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CCGTGTCTGTGCGATGATTGACCGT........................................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................ACAACAACGGACGTGTGTTGACATT.................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CGTGTTCTTTGCAGAGTCGTCGT............................................................................................................................................................................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GTTGACATTAATCAATTGGGTCAATCT................................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GGTATCTTGGGTTTTTATATATATAT.................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................ACACGTTTAACCCCGTGTCTGTGCGAT.................................................................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TTCTGTGCGATGATTGACCGT........................................................................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CCTCGCCGTGTTCTTTGCAGAGTCGT............................................................................................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GGACGTGTGTTGACATTAATCAATT.......................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TCAATCTCTGGTATCTTGGGTTTTTAT............................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CTGGTATCTTGGGTTTTTATATATATAT.................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TGTGCGATGATTGACCGTTGACAT.................................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCTGTGCGATGATTGACCGTTGACAT.................................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TCAATTGGGTCAATCTCTGGTATCT....................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................AATCAATTGGGTCAATCTCTGGTAT......................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TAGTACTACACGTTTAACCCCGTGTCTG........................................................................................................................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................TGTAGAAGGCGCGTCCGG............................................. | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................TGATTGACCGTTGACATCAGGATCAA......................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GACCCTCGCCGTGTTCTTTGCAGAGT.................................................................................................................................................................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CTGGTATCTTGGGTTTTTATATAT........................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TCAACTACTACTACTAGTACTACACGTTT..................................................................................................................................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................CCTGTGTTACTACCACTACTACAACTAGT..................................................................................................................................................................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TGCGATGATTGACCGTTGACATCAGGAT............................................................................................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................GTGTTGACATTAATCAATTGGGTCAAT.................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................AATAAGAACAACAACAACGGACGTGTGT......................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................CAACAACAACGGACGTGTGTTGACAT................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ATCAATTGGGTCAATCTCTAGTATCT....................................................................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TGTGTTGACATTAATCAATTGGGTCAAT.................................................................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GGTCAATCTCTGGTATCTTGGGTTTT............................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................AATCTCTGGTATCTTGGGTTTTTAT............................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................ATGTTGGTAGAAGGTGTCTC................................................ | 20 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................CAAGGAGGTTTGTAGATG....................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TGGCCGTTGACATTAGGGTCA.......................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................CGTTGCCATCAGGATCTA......................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TCAACAACAATAAGAACAACAAC...................................................................................................................................................................................................... | 23 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TCAACAACAATAAGAACAACAACA..................................................................................................................................................................................................... | 24 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ATCAACAACAATAAGAACAACAACA..................................................................................................................................................................................................... | 25 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TCAACAACAATAAGAACAAC......................................................................................................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TCAACAACAATAAGAACAACAA....................................................................................................................................................................................................... | 22 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ATCAACAACAATAAGAACAACA........................................................................................................................................................................................................ | 22 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ATCAACAACAATAAGAACAA.......................................................................................................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................GTTGGTAGAAGGTGTCTCTG.............................................. | 20 | 3 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CCGTGTCTGTCCTATGCTTG............................................................................................................................................................................................................................................. | 20 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACCCTCGCCATGCTCTTAGCA...................................................................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................CTCGGGGACAGGGTACGTGGT....... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................TTGTGCGATGCTTGACCGCTG...................................................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................ATCGGGGGCAGGGTGCGTGTT....... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TTGCCGAGACGTCGCCGCGAC...................................................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................AGGAGGTTTGAAGAAGGCCG................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....CCCCTCGCCGTGCTCTTAGC....................................................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................ATGTAGAAGGCGCGTCCGG............................................. | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........CGGTGGTCTTTGCAGTGTCG................................................................................................................................................................................................................................................................................................................................................................ | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................GGTATCTTGGGCTTTTAG............................................................................................................................................ | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TCAACAACAATAAGAAAAA.......................................................................................................................................................................................................... | 19 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGAAATCTTTGGTATCT....................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................CCAACAACAATAAGAACAACAACA..................................................................................................................................................................................................... | 24 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................GTAGAAGGCTTCTCCGGTTC.......................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:1693882-1694264 - | dvi_3436 | CGTCTGGGAGCGGCACAAGAAACGTCTCAGCAGCAGCGCTGACAACCAGGGTGAGGCGGGTGGACACAATGA------------------------------------------------TGGTGA------------------------TGATG---------TTGATCATG---AAGTTGATGATGATGATCATGATGTGCAAATTGGGGCACAGACA----------CGCTACTAAC--T-----GGCAA-CTGTAGTCCTAGTTGTTGTTATTCTTG---TTGTT------GTTGCCTGCACAC-----------AA-------------------------------------CTGTAATTAGT----TAACCCAG-----TTAGAGACCA-TAGAACCCAAAAATATATATATATATATATAAACAAAAAAGTTCACTT------------------------------------------------------------------------------------------------GC---------AATTC-A--------ACTGAACCCA---A-----------CTG-----------------AACTCAACTGAACTTC----CTCCAAACATCTTCCGCAGAGGCCCGGAGCTATGCTGA-CTTTAGCCCCAGTCCCATGCACGATGCTTCC |
| droMoj3 | scaffold_6540:27412919-27413264 - | CGTCTGGGAGCGGCACAAGAAGCGCCTCAA---CAGCGCTGACAACCAGGGTGAGGCGGGTGAACACC---ATG---A------------------------------------------TCATGCTCA---------------------TAATGA------------------------TGATGATGATGATAATGATGTGCAAATTGGGGCACAGACA----------CGTTACTAACG-------GGCAA-CTGTAGTC------CTAG---TTGTTG---TTGAT------ATTGCCTGCACAC-----------AG-------------------------------------CTGTAATTAGT----TAACCCAG----TTTAGAGACCA-TTAAA-CCA---------------------------AATAACCCACTTGCAAT-------------------------------------------------------------------------------------------CC---------AATCGAA-TCC--CCACTGAACTCA---A-----------CTG-----------------AAATCAACTGAACTTC----CTCCACACATCTTCCGCAGAGGCCCGGAGCTATGCTGA-CTTTAGCCCCAGTCCCATGCACGATGCATCC | |
| droGri2 | scaffold_14624:4228127-4228462 - | CGTCTGGGAGCGGCACAAGAAACGTTTGGGCAGCAGCGCTGACAACCAGGGTGAGACGGGTGGACACA------------A---------------------------------------TGACAATGA---------------------CAATGA------------------------CAATGATGATGACGCTGATGTGCAAATTGGGGCACAAACA----------CGTTACTAACC-T-----GGCAA-CTGTAGTC------CTTG------------TTGTT------GTTGCATGCACAC-----------AG-------------------------------------CTGTAATTAGAAATCTAACCCC-AAAAGATAGAGAGAAATAGAACCCA---------------------------AAT-ATCCACTT--------------------------------------------------------------------------------------------------------------GC-A--------GCTACAGCTACACT-----------T-----------------ACAACTCAACTGAACTTC----CTCCAACCATCTTCCGCAGAGGCCCGCAGCCATGCAGA-CTTTAACCCCAGTCAGATGCACGATGCATCC | |
| droWil2 | scf2_1100000004943:9043293-9043686 - | CGTCTGGGAGCGGCACAAGAAACGTTTGGG---CAGCGCCGGCAATGAGGGTGAGGCGGGTGGAGATC---AAA---AAAA---------------------------------------TG---ATGA---------------------TAATGA------------------------TGATGAAGACGATAATGACGTGCAATTTGGGGCAAAGACA----------CGTTACTAA---------GGCAA-CTGTAGTC------GTTGTTA--ATTG---TTGTTATTATTATTGCATGCACAATATATACATATATAT--ATACCTACC-----------CCCGATACCTGAACCGTATTTAAATATCTAGTCTA-C----------------------------------------------AATAACATCGTAGAGTTGCAATCCTCTAGCAGAAAACTGAACTT-CTTAACC----------------AAAGCAAA-----------------------------A-CA--------------------------------------------------------ACAAAT-----CA-AT-TTGAATGAACTCTACTTC----CTCAACTAATCTTCCGCAGAGGCCCT-------GCTGATCTATAGCCCCAGTCCAATGCACGATCCCTCG | |
| dp5 | 2:837076-837486 + | GGTCTGGGAGCGACACAAGAAACGCTTGGC-TG--CCGCCGAAAGTCAGGGTGAGGCGGGAGGAGACAAACACG---G---TCGTAC------T---C------GT---AATAGCAATGATAAAGGCGATAAAGACGATAAAGA------CGATA---------ACGATAACG---ATAACGATGATGATGAAAGTGACGTTCGATTTGGGGCGAAGTCACAACAGTC-CCGCTACTAACGATGCCTGCTCGTACCGTAGTT------GTTGCTTTAATTT---ATTTA------GTTAGTTGTAG--------------ACACGATTA----------------CTTGATGA---------------------------------------------------------------------------------------------------------------------------------------------ACAACGTACATACCT--CTTAGCAAGAGCCTTCCC-----------CCC---------TTTCC-A--------ACTCGAGCCACACA-----------C----------ACA--TTCCAACTGAACTTAAATTC----TCCAATCAATCTTCCCCAGAGGCTCTGGCCTATGCAGA-CTATAACGCAAGTCCTTTTCACAATCCCTCG | |
| droPer2 | scaffold_7:3619510-3619914 + | GGTCTGGGAGCGACACAAGAAACGCTTGGC-TG--CCGCCGAAAGTCAGGGTGAGGCGGGAGGAGACAAACACG---G---TCATAC------T---C------GT---AATAGCGATGATAAAGACGA---------------------TGAAGACGATAAAGACGATAACG---ATAACGATGATGATGAAAGTGACGTTCGATTTGGGGCGAAGTCACAACAGTC-CCGCTACTAACGATGCCTGCTCGTACCGTAGTT------GTTGCTTTAATTT---ATTTA------GTTAGTTGTAG--------------ACACGATTA----------------CTTGATGA---------------------------------------------------------------------------------------------------------------------------------------------ACAACGTAGATACCT--CTTAGCAAGAGCCTTCCC-----------CCC---------TTTCC-A--------ACTCGAGCCACACA-----------C----------ACA--TTCCAACTGAACTTAAATTC----TCCAATCAATCTTCCCCAGAGGCTCTGGCCTATGCAGA-CTATAACGCAAGTCCTTTTCACAATCCCTCG | |
| droAna3 | scaffold_13340:20732260-20732657 + | dan_3200 | GGTCTGGGAGCGTCACAAGAAAA--------AACACAGC-------CAGGGTGAGGCGGGTGGAAAAAAATCGA---A---TCCTAA------TAACCACAATA---ACC---ACGATAACAAGAACAA---------------------GGACGAGGGCCAGGACGACGACG---AGGAGGACGACGACAATGGGGACGTGCGATTTGGGGAGAAGTCG----AGTCCCCGTTACTAG---------GGCAA-CCGTAGTC------GTTGATTTTATTA---TTGTTATTATAGT-TGTTGCACCC---------------------------------------CA-------AAAA---------------------------------------------------------------------------------------------------CCCAACTAGACTTGATTAACC----------------AAAGTAGA-----------------------------A-CTAGTCCC---------AGTCCCG----TACGAACC------------GACCACAATGCCAGGTCAGCATG-TT-TTCATTGAGTTCTCATCA----TCCCATCAATCTTTCCCAGAGGCCCTGACCTACGCGGA-CTATAATGCCAGTCCGGTGCACAATCCCTCG |
| droBip1 | scf7180000396413:1665627-1666023 - | GGTCTGGGAGAGGCACAAGAAGA--------AGCACAGC-------CAGGGTGAGGCGGGTGGAAAAAAATCGA---A---TCCTAA------TAACCACAATA---ACC---ACGATAACAAGAACAA---------------------GGACGAGGGCCAGGACGACGACG---AGGAGGACGACGACAACGGGGACGTGCGATTTGGGGAGAAGTCG----AGTCCCCGTTACTAG---------GGCAA-CTGTAGTC------GTTGATTTTATTA---TTGTTATTATTGT-TGTTGCACCA-----------A------------------------------------AAAA---------------------------------------------------------------------------------------------------CCAAACTAGACTTGATTAACC----------------AAAGTAGA-----------------------------A-CTAGTCCC---------AGTCCCG----TACCAACC------------GACCACAATGCCAGGTCAGCATG-TT-TTCATTGAGTTCTCATCA----TCCCATCAATCTTTCCCAGAGGCCCTGACCTACGCGGA-CTATAATGCCAGTCCGGTGCACAATCCTTCG | |
| droKik1 | scf7180000302475:1785300-1785697 + | CGTGTGGGAGCGGCACAAGAAGA--------AGCAGAGC-------CAGGGTGAGCCGGGTGGCGGAAATCCAA---A---TCCCAA------T---C------CCAGTC---CCAGTAACAACGACAA---------------------CGACGACGGCAACGACGCCG------CCCCCGATGAGGACGATGGCGACGTGCGTTTCGGGGCCAAGTCG----------CGTTACTAG---------GACAACCCGTAGTC------GTTGATTAACTAA---CTGTT------ATTGCTTGCACAC-----------AC-CAG---A----------------C------------CCGTAACTAACAACG---------------------------------------------------------------------------------------------------------------TAGCT-AATAGCGA--TTGCGACCTAT-----------------G--TTACCT-CT-CCACCACCTGTAGTCC--------------------CTCGCCATCT---CGGTAAAGGCCGCTCC-CT-TATCCCGTACTCAACCTCTCTCTCGAATCGATCTTCCCCAGAGGCCCTGACCTACGCGGA-CTACCACGCGAGTCCGCTGCACAATCCCTCG | |
| droFic1 | scf7180000453826:865070-865418 - | GGTCTGGGAGCGGCACAAGAAGA--------AGCAGACT-------CAGGGTGAGCCGGAAAAAGAAACCCCCAA--A-------------------CCCGAAA---GTC---CTAACAATAATAATAA---------------------CGATGA---------------------------------CGACGAGGACGTGCGATTCGGGGCCAAGTCG----------CGTTACTAAG--------GACAACCTGTAGTC------GTTGATTTAGTTGTTATTGTT------ATTGCTTGCACCA-----------ATCCTGATTA----------------CCCGATAACCAAACCG---------------------------------------------------------------------------------------------------------------------------TAGCTAACTAACAACGTAGACACCT--------------------------------CC---------ACT--------------------------TCAACC---TGGTAAAGCCACTCCA-CT-TTTATCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCTCGACTATGCGGA-CTATGATCCCAGTCCGCTGCACAATCCGTCG | |
| droEle1 | scf7180000491280:2002249-2002619 + | GGTCTGGGAGCGGCACAAGAAGA--------AGCAGTCA-------CAGGGTGAGGCGGGCGGAGAAACGACAAAAAAGAATAAGAA------TAAGA------A---TA---ATAATAATAATAATAA------------CAATGATAACGATGA------------------------CGACGAGGACGATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTACTAG---------GACAACCTGTAGTC------GTTGATTTTGTAA---TTGTT------ATTGCTAGCACCA-----------ATCCTGATAA----------------CCCGATAACCAAAAA---------------------------------------------------------------------------------------------------CCAA-------------AACCAAAGTAGCTAACTAACAGCATAGAGACCTAC-----------------C-----------CGC---------AAACC-C--------ACT--------------------------------------TAACACTTGTACTCAACCTC----TCCCATCGATCTTCCCCAGAGGCCCTGACCTACGCGGA-CTATAATCCCAGTCCGCTGCACAATCCGTCG | |
| droRho1 | scf7180000779336:251560-251925 - | GGTCTGGGAGCGGCACAAGAAGA--------AGCAGTCGTCA----CAGGGTGAGGCGGGCGGAGAAACCCCAA---ATAA---TAA------T---------------------AATAATAATAATAA---------------------CGATGA------------------------CGACGAGGACGAAGGCGACGTTCGATTTGGGGCCAGGTCG----------CGTTACTAG---------GACAACCTGTAGTC------GTTGATTTAGT-A---TTGTT------ATTGCTTGCACCC-----------ATCCTGATAA----------------CCCGATAACCAAAATG---------------------------------------------------------------------------------------------------------------------------TAGCTAACTAACAACGTAGAGACCTAC-----------------C-----------TCC---------AGTCC-CACCA--CCGGTG------------AACC---CGGTCAAGTCAGCTAC-CTAACAATTGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCTGACCTATGCCGA-TTATAATCCCAGTCCGCTGCACAATCCGTCG | |
| droBia1 | scf7180000302113:1670572-1670963 - | GGTCTGGGAGCGGCACAAGAAGA--------AGCAGTCG-------CAGGGTGAGCCGGGAGGAGAAACCCCAA---AGAATAAAAC------TAACC------G---TA---ACAGTAATAATAATAA---------------------CGATGACGACGAGGACGAGG------ACGAGGACGATGACAATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTAGTAG---------GACAACCTGTAGTC------GTTGATTTTGTTA---TTGTT------ATTGCTTGCACCACAAATCCCGACATCCTGATAA-------------CATCCCGATAACCAAACCG---------------------------------------------------------------------------------------------------------------------------TAGCCAACTAACAACGTAGTCACAC-C-----------------CTCT-------------------------------------------------TCAACC---TGGTAATGTCAGTCCC-CC-TATATCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCTCGACTATGCGGA-CTACAATCCCAGTCCGCTGCACAATCCGTCG | |
| droTak1 | scf7180000415765:112730-113138 + | CGTCTGGGAGCGGCACAAGAAGA--------AGCAGACG-------CAGGGTGAGCCGGGAGGAGAAACCCCAA---AGAATACAAATAAAACTAACCACAAAA---ATA---ATAATAATAATAATAA---------------------CGATGAC------------GACG---ACGAGGACGATGACAATGGCGACGTTCAGTTTGGGGCCAAGTCG----------CGTTACTAAG--------GACAACCTGTAGTC------GTTGATTTTGTTA---TTGTT------ATTGCTTGCACCAAAAATCCTGACATCCTGATAGC--CCCCCGATAACATCCCGATAACCAAAACG---------------------------------------------------------------------------------------------------------------------------TAGCCAACTAACAACGTAGATACAC-C-----------------CTCT---------CT--------------------------------------GCAAAC---TGGTAAAGTTAGTCAC-CT-TTTACCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCTGAACTATGGTGA-TTACAATCCCAGTCCACTGCACAATCCGTCG | |
| droEug1 | scf7180000409555:208399-208778 + | GGTCTGGGAGCGGCACAAGAAGA--------AGCAGTCA-------CAGGGTGAGCCGGGAGGAGAAACCCCAA---AGAA---------------------------------------TAACAATAA---------------------CGATGAC------------GACGAGGACGACGACAATGACGATGGCGATGTTCGATTTGGGGACAAGTCG----------CGTTACTAG---------GACAACCTGTAGTC------GTTGATTTTGTTA---TTGTT------ATTGCTTGCACCA-----------ATCCTGATAA----------------CCCGATAACCAAAACG---------------------------------------------------------------------------------------------------------------------------TAGCTAACTAACAACGTAGAGACCTAC-----------------CTCTAA-------CC---------AGTCCAGTCCAGTTCAGTC-----GCACTTCAACC---TGGTAAAGTCAGTCCT-CT-TATAATGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCTGACCTACACGGA-CTATAATCCCAGTCCGCTGCACAATCCGTCG | |
| dm3 | chr3R:25737635-25737993 + | GGTCTGGGAGCGGCACAAGAAGA--------AACAGACG-------CAGGGTGAGCCGGGCGGAGAAACCCCAA---AGAG---------------------------------------TAACAATAA---------------------CGATGA------------------------CGATGAGGACGATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTACTAG---------GACAACCCGTAGTC------GTTGATTTAGTTA---TTGTT------ATTGCTTGCACCA-----------ATCCTGATAA----------------CCCGATAACCAAAACG---------------------------------------------------------------------------------------------------------------------------TAGCT-ACTAACAACGTAGAGACCCAC-----------------CTCT---------CC---------AGTCCTCTCCATATCAGAC------------GACC---TGGTAAAGTCATTCCT-CCGCCTATCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCAGACCTATGCCGA-CTACAACCCCAGTCCGCTGCACAATCCGTCG | |
| droSim2 | 3r:25083795-25084153 + | dsi_17220 | GGTCTGGGAGCGACACAAGAAGA--------AACAGACG-------CAGGGTGAGCCGGGCGGAGAAACCCCAA---AGAG---------------------------------------TAACAATAA---------------------CGATGA------------------------CGATGAGGACGATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTACTAG---------GACAACCCGTAGTC------GTTGATATAGTTA---TTGTT------ATTGCTTGCACCA-----------ATCCTGATAA----------------CCCGATAACCAAAACG---------------------------------------------------------------------------------------------------------------------------TAGCT-ACTAACAACGTAGAGACCCAC-----------------CTCT---------CC---------AGTCCTCTCCATATCAGTC------------GACC---TGGTAAAGTCATTCCC-CCGCCTATCATACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCAATCCTATGCCGA-CTACAACCCCAGTCCGCTGCACAATCCGTCG |
| droSec2 | scaffold_4:4579307-4579665 + | dse_1298 | GGTCTGGGAGCGGCACAAAAAGA--------AACAGACG-------CAGGGTGAGCCGGGCGGAGAAACCCCAA---AAAG---------------------------------------TAACAATAA---------------------CGATGA------------------------CGATGAGGACGATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTACTAG---------GACAACCCGTAGTC------GTTGATATAGTTA---TTGTT------ATTGCTTGCACCA-----------ATCCTGATAA----------------CCCGATAACCATAGCG---------------------------------------------------------------------------------------------------------------------------TAGCT-ACTAACAACGTAGATACCCAC-----------------CTCT---------CC---------AGTCCTCTCCATATCAGTC------------GACC---TGGTAAAGTCATTCCC-CCGCCTATCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCAGTCCTATGCCGA-CTACAACCCCAGTCCGCTGCACAATCCGTCG |
| droYak3 | 3R:20735445-20735816 - | GGTCTGGGAGCGGCACAAGAAGA--------AACAGACG-------CAGGGTGAGCCGGGCGGAGAAACCCCAAAAAAGAG---------------------------------------TAACAATAA---------------------CGATGAC------------GATG---AGGACGATAAGGACGATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTACTAG---------GACAACCCGTAGTC------GTTGATTTAGTTA---TTGTT------ATTGCTTGCACCA-----------GTCCTGATAA----------------CCCGATAACCAAAACG---------------------------------------------------------------------------------------------------------------------------TAGCT-ACTAACAACGTAGAGACCCAC-----------------CTCT---------CC---------ACTCCTCTCCATATCAGAC------------GACC---TGGTAAAGTCATTCCCCCCGCCTCTCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCACACCTATGCCGA-CTACAATCCCAGTCCGCTGCACAATCCGTCG | |
| droEre2 | scaffold_4820:2270897-2271255 - | GGTCTGGGAGCGGCACAAGAAGA--------AACAGACG-------CAGGGTGAGCCGGGCGGAGAAAACCCAA---AGAG---------------------------------------TAACAATAG---------------------CGATGA------------------------CGATGATGACGATGGCGACGTTCGATTTGGGGCCAAGTCG----------CGTTACTAG---------GACAACCCGTAGTC------GTTGATTTAGTTA---TTGTT------ATTGCTTGCACCA-----------GTCCTGATAA----------------CCCGATAACCAAAACG---------------------------------------------------------------------------------------------------------------------------TAGCT-ACTAACAACGTAGAGACCCAC-----------------CTCT---------CC---------AGTCCTCTCCATATCAGTC------------GACC---TGGTAAAGTCATTCCC-TCGCCTATCGTACTCAACTTC----TCCAATCGATCTTCCCCAGAGGCCCAGACCTATGCCGA-CTACAACCCCAGTCCGCTGCACAATCCGTCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:24 AM