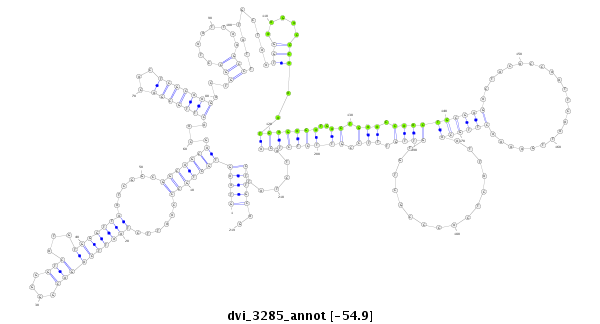
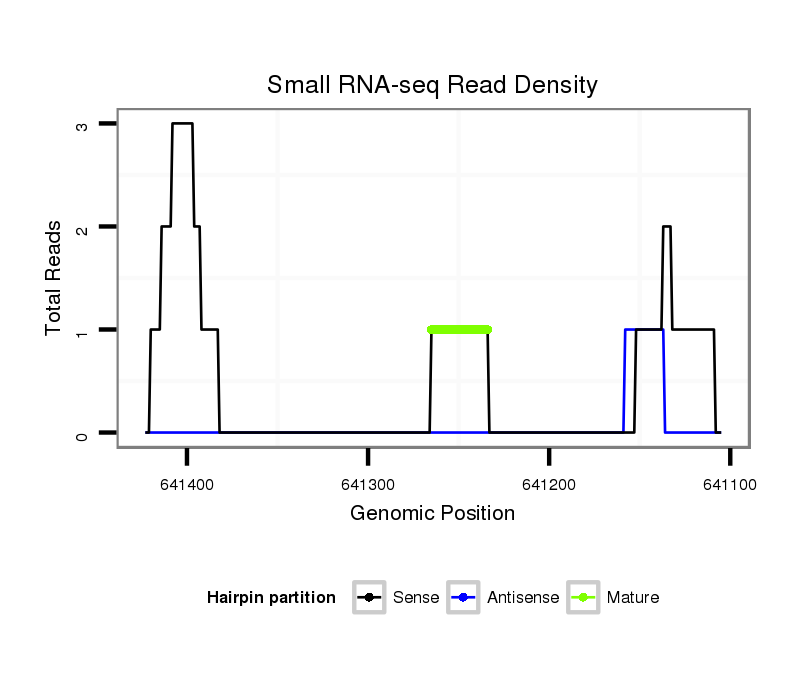
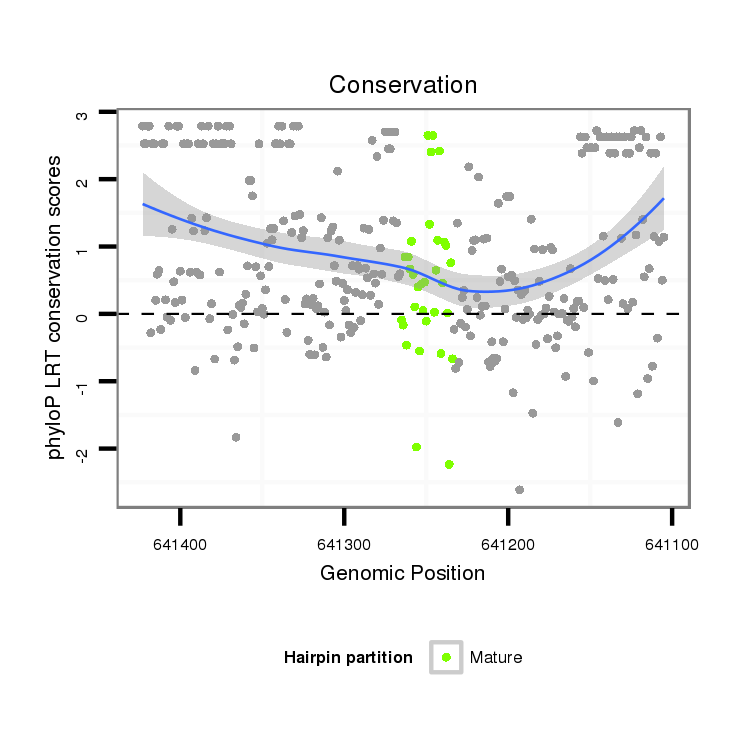
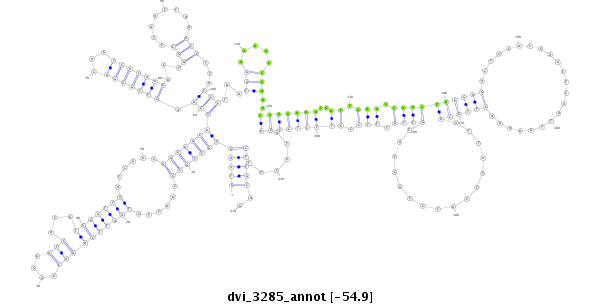
ID:dvi_3285 |
Coordinate:scaffold_12855:641155-641373 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -54.9 | -54.9 | -54.9 |
 |
 |
 |
CDS [Dvir\GJ10557-cds]; exon [dvir_GLEANR_10473:1]; exon [dvir_GLEANR_10473:2]; CDS [Dvir\GJ10557-cds]; intron [Dvir\GJ10557-in]
No Repeatable elements found
| ##################################################---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCATTGGCAAGGGTAAACGGAAGGCTGGCACCACAACCATTAGACAGCGGTAAGTGCTGCCCAATTCTAATTCAGACGGGGGTCATGTGGGTTAATCGGCGGCAGCAGCAACTTGCGACACTCGCAAGACTGGCCTAATTAAGCCTTATGCTAATGGAAGACCCAAATGAGAGATGACTGAACCAACTGCGAGACTACACCAATTCAATTGAAAACTTGCAATTACTCACCCGCTGTGTTGTTTCCGTTCTCTCAATCTATCTTGCAGCTCTGGTGACATGGTCGTGGTGACCACCGAGTCCGAACTGACACCCGACA **************************************************((((((((((((......((((((((((....)))...)))))))......)))))))....(((((((...)))))))...(((........))).........(((.....)))..(((((((..((.(((.((((((((((......................))))))................)))).))).)))))))))......)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR1106720 embryo_8-10h |
SRR060655 9x160_testes_total |
SRR060657 140_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106715 embryo_4-6h |
V116 male body |
M047 female body |
M061 embryo |
SRR060672 9x160_females_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
V047 embryo |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................GGTGACATGATCGTGGTGACCACCGA.................... | 26 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........AAGGGTAAACGGAAGGCT.................................................................................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ATTGGCAAGGGTAAACGGAAGGCTGGCA................................................................................................................................................................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TCATGTGGGTTGAACGGCGGC....................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AAACGGAAGGCTGGCACCACAACCAT...................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TACCCAATCCTAATTCAGA.................................................................................................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................TGGTGACCACCGAGTCCGAACTGACACCCGAT. | 32 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................CTGGTGACATGGTCGTGGTG............................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGACCCAAATGAGAGATGACTGAACCAACTG................................................................................................................................. | 32 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................TGGTGACCACCGAGTCCGAACTGACACCC.... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TACCCAATCCTAATTCAG................................................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.86 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| ..........................................................TACCCAATTCTAATTCAGG.................................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CTACCCAATCCTAATTCAGG.................................................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCCCAATCCTAATTCAGG.................................................................................................................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCCAATCCTAATTCAGAGAG............................................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................CTTCGACCCTCGCAAGACT........................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........TGGTGACCGGAAGGCTGGC................................................................................................................................................................................................................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................ATGGAAGACCCCATTGAGG.................................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................GTTGTTGCCGGTCTCTCA............................................................... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ACGTAACCGTTCCCATTTGCCTTCCGACCGTGGTGTTGGTAATCTGTCGCCATTCACGACGGGTTAAGATTAAGTCTGCCCCCAGTACACCCAATTAGCCGCCGTCGTCGTTGAACGCTGTGAGCGTTCTGACCGGATTAATTCGGAATACGATTACCTTCTGGGTTTACTCTCTACTGACTTGGTTGACGCTCTGATGTGGTTAAGTTAACTTTTGAACGTTAATGAGTGGGCGACACAACAAAGGCAAGAGAGTTAGATAGAACGTCGAGACCACTGTACCAGCACCACTGGTGGCTCAGGCTTGACTGTGGGCTGT
**************************************************((((((((((((......((((((((((....)))...)))))))......)))))))....(((((((...)))))))...(((........))).........(((.....)))..(((((((..((.(((.((((((((((......................))))))................)))).))).)))))))))......)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060677 Argx9_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
M027 male body |
SRR060657 140_testes_total |
SRR060666 160_males_carcasses_total |
V053 head |
SRR060680 9xArg_testes_total |
M047 female body |
SRR060681 Argx9_testes_total |
V116 male body |
SRR060679 140x9_testes_total |
GSM1528803 follicle cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................................GAGACCAATGTACCAGCACC.............................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................CGTCGAGACCACTGTACCAGCA................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................CTGGCTTGACTGAGTGCTG. | 19 | 3 | 6 | 0.33 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................CAGCACAACTGGTGGCTTAA................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................GGGCTAGACTGTGCGCTGT | 19 | 3 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................CTGTACCGGAACCACTGTTG....................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................ATGGGTTAGGATTAAGTC................................................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTCCGGGTTTGCTCTCTA............................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................CTGATTGGGCGACACAAGA............................................................................ | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................GGCGACACACCAAAGACA...................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................ACCGTGGAATTGGTAATCC.................................................................................................................................................................................................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................CCCTGTACACCCAGTTAGT............................................................................................................................................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................................ACCGGGGGCTCAGGCTAG............ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12855:641105-641423 - | dvi_3285 | TGCATTGGCAAGGGTAAACGGAAGGCTGGCACCACAACCATTAGACAGCGGTAAGTGCT--G-----------------------------------CCCA-ATTCTAATTCAGA------------CGGGGGTCATG---TGGGTTAATCGGCGGCAGCAGCAACTTG--CGACACTCGCA------AGA---------------CTGG-CCTAATTA----AGCCTTATGCTAATGGAAGACC------------CAAATG-AGAGATGACTGAACC-------------AACTG--CGAG---AC--TACACCAA-----------------------------------------------------------------------------------------TTCAATTG------------------------------------------------------AAAACT-----TG--------------------------------------------------------CAA---------TTACTCACCC-----GCTGTGTT------------GTT------TC-------CGTTCTCTCAATCTAT-CTTGCAGCTCTGGTGACATGGTCGTGGTGACCACCGAGTCCGAACTGACACCCGACA |
| droMoj3 | scaffold_6540:12329540-12329862 + | dmo_1914 | TGCATCGGCAAGGGACAGCGCAAGGCAGGCACCACAACAATTAGGCAACGGTAAGTGCC--G-----------------------------------CACA-A------TTCTAA-------GCAGACGGAGGTCATG---CGGGTTAATCGGCAGACACAGCGTCTTG--CGACACTCGCA------AGG---------------CTGG-CCTAATTA----AGCCTTATGCTAATGGGTGCC--G----------CAAATG-GAAGATGAGCGAAGC------------AAA-TG--CGAG---ACTTTGCACCAA-----------------------------------------------------------------------------------------TTCAATTA------------------------------------------------------AATTTTTAACGTG----T---------------------------------------------------TTA---------TTACTCATGC-----GTTGTTTT------------GTT------TC-------CGCTCTTC---CGCCT-CTTGCAGCTCCGGCGACATGGTTGTGGTCACCACGGAGTCCGAACTATCTTCAGACG |
| droGri2 | scaffold_14906:1750452-1750750 - | TGCATTGGCAAAGGCGAACGCAAATCAGGCACCACAACCCTCAGGCAGCGGTGAGTCGG-AG-----------------------------------TCCA-ATTCTAATTCAGA------------CGGGGGTCATT---GGGGTTAATCGGCGAGAACAGCATATTG--CGACACTCACA----------------C-TTGAGACTGG-CCTAATTA----AGCCTTATGCTAATGGAA-----------------------------------CGCTGA---------CAAAATGAC------------CACCAT-----------------------------------------------------------------------------------------TTCAATTA------------------------------------------------------ACTT--------GCTTGC---------------------------------------------------CAA---------TTAGTCAGCCAGTTCATCTTC-AT---------------C--------TTCCG--C--------TTCC-ACTTTCAGCTCCGGCGACATGATTGTGGTAACTACGGAATCGGATCTGACACCCGATG | |
| droWil2 | scf2_1100000004902:5559110-5559394 - | TGCATAGGACGGGGCAAACCCAAGGCTGGCACAGCGACCATTAGACAGCGGTAAGTAAA-TG-----------------------------------ATCTTTTTCAATTTTC------G--TCGTCTAGGGGTCAAG---TGGTTAAATGGCTCAGA--------------CACAGACACCCACACTC----CCT-------------A-ACTAATTAGAGCGGTGTTATGCTAATAGA--------------ATCCATATT-GCTGAAGATC-------------------------------------CAAATGA-----------------------------------------------------------------------------------------TTCAATTA------------------------------------------------------AAGTTTC---------GC---------------------------------------------------CAA---------TTACTCAT---------------A---------------C--------TTCCG--T--------TTCGT-CTTACAGCTCTGGCGATATGATTGTTGTGACCACCGAATCGGAATTGACACCCGATG | |
| dp5 | 2:2179741-2180030 - | TGCATCGGGAAGGGTCAGCGCAAATCGGGGCACGCAACCATTCGACAGCGGTGAGTGG---------------------------------------TCTA-ATTCGA----------GT--GTGTCTGGGGGTCAAG---TGGGTTAATCGGCGGCAATAGCT---CGCCCCACACT----------C--------------------C-GCTAATTA----GGTCTTATGCTAATGGAAGCCCCGCAGATGCTTCCTCCTG-GCGTATGACTAACGCCGA---------GAATTTGAT--ACTT---------------------------------------------------------------------T-------------------------------TTCAATTA------------------------------------------------------GTTTCT--------------------------------------------------------------------------------TAA---------------A---------------A--------TTTCGTTC----C---CCCT-A--CACAGATCCGGCGACATGATTGTGGTCACCACCGAGTCCGAATTGACGCCCGATG | |
| droPer2 | scaffold_7:2347238-2347527 - | TGCATCGGGAAGGGTCAGAGAAAATCGGGCCACGCAACCATTCGACAGCGGTGAGTGG---------------------------------------TCTA-ATTCGA----------GT--GTGTCTGGGGGTCAAG---TGGGTTAATCGGCGGCAATAGCT---CGCCCCACACT----------C--------------------C-GCTAATTA----GGTCTTATGCTAATGGAAGCCCCGCAGATGCTTCCTCCTG-GCGTATGACTAACGCCGA---------GAATTTGAT--ACTT---------------------------------------------------------------------T-------------------------------TTCAATTA------------------------------------------------------GTTTCT--------------------------------------------------------------------------------TAA---------------A---------------A--------TTTCGTTC----C---CCCT-A--CACAGATCCGGCGACATGATTGTGGTCACCACCGAGTCCGAATTGACGCCCGATG | |
| droAna3 | scaffold_12911:5006488-5006797 - | TGCATCGGGCGGGGCGAGCCCAAGGCGGGCACAGCCACCATCAGACAGCGGTAAGTGGTTCG-----------------------------CAGAATTCTA-ATT----TTACGC-TCGGACGTGTCTGGGGGTCAAG---TGGGTTAATCGGATAGACTG------------------GGC------TGCTCCTCAAC-GTGGGGA--GCTTTAATTA----TGCCGTATGCTAAC--T--------------CACAATGCCATGGAATGAGTAATGCAAC---------CAT-----------------CAATTAATACATTTTAAA---------------------------------------------------------------------------------------------------------------------AAAAAAAAAAACAAATTTAATTAAATA-------------------------------------------------------------------------------------------------------------------------------ATA-----------TTAT-TATTACAGCTCTGGTGACATGATTGTGGTTACAACTGAATCCGAACTAACTCCTGATG | |
| droBip1 | scf7180000396640:1842989-1843311 - | TGCATCGGGCGGGGCAAGCCCAAGGCGGGCACCGCCACCATCCGACAACGGTAAGTGGTTCG-----------------------------CGAAACTCGA-ATTCTACT-CGGACTCGGACGTGTCTGGGGGTCAAG---TGGGTTAATCTGATAGACTGGCGTGTT---------------------G---CTCAAAGTTGGGAG--C-TTCAATTA----GGTCGTATGCTAATGAT--------------GAAAGCTGT-GGGGATGAGTAACGCAGT---------CATA-----------------AATTAATA--------------------------------------------------------TACGTAT--------TTTTCGGGAA---------------------------------------------ATATTATATAGAGAATTCAATATTA------------------------------------------------------------------------------------------------------T---------------A--------TTA-------------TAATT-TTTACAGCTCTGGTGACATGATTGTGGTAACAACTGAATCCGAACTAACTCCTGATG | |
| droKik1 | scf7180000302466:111328-111750 + | TGCATCGGGCGGGGCAAGCCCAAGACTGGCACGGCCACCATCAGTCAGCGGTGAGTGGT-CACAGGGGGAGGTGTATGGTATCCGAGATCCGGACATTCTA-ATTCCAATTCCGACGCGACTGTCTCTGGGGGTCAAG---TGGGTTAATCGATTGGTTAACCG---GG----ACACACATT------CACTCT-CAAG-ATGAGAC--G-TCTAATTA----GGCCTTATGCTAATAGG--------------GACCACGCT-GCGTATGAGTAATAACAT---T---------------------G--CCAAATAAGACAATTCAGAGT-------AGTTGAGATCGGTAAAAT------------------------------------------------------AATTGATACATTT-------------------------------------------------------------------GTAGGGTTCTTCTTTAGCTTAACTTTAAGTTTTAATATTAATTTACTTTAA---------ATAT-TTA---------------A---------------A--------TTTTAAT---------TTTTT-TCAACAGATCCGGTGAAATAATTGTGGTAAATACCGAGTCGGAACTCTCACCCCATG | |
| droFic1 | scf7180000453776:618562-618907 - | TGCATCGGGCGTGGCAAGCCGAAGGCGGGAACAACCACCATCCGGCAGCGGTAAGTAC---------------------------------------TCTA-A------TTCCGA-TCGA--GTGTCTG-GGGTCAAG---TGGGTTAAACGGCTTGC--------------CGCGTTTGTT------TGCTCGCCAAC-ATGGGGC--G-TCTGGTTT----GGCCTTATGCTAATGAT--------------GACCACTCT-GCGGATGAGTAACGCA----------------------------------ATAAAACAATTAAGAAACCATAAAACTT-------------TCTTAA---------------TACGTATGGATCATATTTT-GGGGATTGGGA--AGATTG------------------------------------------------------G-TT-------------------------------------------------------------------------------------CTAAAACGA--------------------AACTAAGTTCGTT---TTC----T--TTCTTT-TTAATAGCTCTGGAGACATGGTTGTGGTCACCACCGAGTCCGAACTTACTCCCGATG | |
| droEle1 | scf7180000491280:1533581-1533807 - | TGCATTGGGCGTGGCAAGCCCAAGGCGGGCACAGCCACCATCCGGCAGCGGTGAGACT---------------------------------------TCTA-A------TTCCGA-TCGG--GTGTCTG-GGGTCAAG---TGGGTTAATCGGCTTGC--------------CACATTTGTT------TGCTCGCCAAC-ATGGGGC--G-TCTGGTTT----GGCCTTATGCTAATGAT--------------GACCACTCT-GCGGATGAGTAACGCGGC---------AATATGTGCCAG---AC--CCCATTAAAACAATTTAAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCGGCGA----------------------------------------- | |
| droRho1 | scf7180000777929:21398-21625 - | TGCATCGGGCGTGGCAAACCCAAGGCGGGCACAGCCACCATCCGGCAGCGGTAAGTAT---------------------------------------TCTA-A------TTCCGA-TCGA--GTGTCTGGGGGTCAAG---CGGGTTAATCGGCTTGC--------------CACATTTGTT------TGCTCGCCATC-ATGGGGC--G-TCTGGGTT----GGCCTTATGCTAATGAT--------------GACCACTCT-GCGGATGAGTAACGCAGC---------AATATGTGCCAG---GC--CTCAATAAATCAATTAAAAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGGGAA------------- | |
| droBia1 | scf7180000302136:2789128-2789513 + | TGCATCGGGCGTGGCAAGCCCAAGGCGGGCACAGCCACCATCCGGCAGCGGTGAGAGCT---------------------------------------CCC-CTTCCCATTCCGA-TCGA--GTGTCTGGGGGTCAACTGGGGGGTTAATCGGCTTGC--------------CGCACTTGTT------TGCCCGCCAGC-ATGGGGC--G-TCCAGTTT----GGCCTTATGCTAATGAT--------------GACCACCCG-GCGGATGAGTAACGCAGC---------AAATTGCGAAAT---GC--CCCATTAAAACAATTAAGCACTAG-G-AAGAT-------------TCTTTAAACAAATGGT------------------------------------GTTGAGAA---------------------------------------------------------AA----------AGTAGTGTGGGATTCTTAAATAGA----------------------------------------------TCAGCT-----GACTTATTATTAATTATATTT---T--------TTATG--C--------TTTAC-CCAACAGTTCTGGCGACATGATTGTGGTGACCACCGAGTCCGAACTCACACCTGATG | |
| droTak1 | scf7180000415765:859789-860154 + | TGCATCGGGCGGGGCAAACCCAAGGCGGGCACAGCCACCATCCGGCAGCGGTAAGAGTT-CC--------------------------------AATTCCA-ATTCCAATTCCGA-TCGAGTGTGTCTGGGGGTCAAGTG-GGGGTTAATCGGCTTGC---------------------------------TCGCCAAC-ATGGGGC--C-TCTAGTTT----GGCCTTATGCTAATGAT--------------GACCACGCT-GCGGATGAGTAATGCA----------------------------------ATAAAACAATTAAGAACTGGAG-AAGTT-------------T------------------------------------------------------ACTTGAAAAATTTAGAGTGGTTAGGGGTAATTTAAAAGAAATA--------------------------------------------------------------------ATAAGGGTAACCTACCT-AAATATATATTTTAATTATCC-----GACTTA---------------------------------------T---TTTTC-CCTATAGTTCTGGCGACATGATCGTGGTGACCACCGAGTCTGAACTTACACCAGATG | |
| droEug1 | scf7180000409787:243644-243991 + | TGCATCGGGCGTGGCAAGCCCAAGGCGGGCACGGCCACCATCCGGCAGCGGTAAGCAT---------------------------------------TCTA-A------TTCCGA-TCTG--GTGTCTGGGGGTCAAG---TGGGTTAATCGGCATGC---------------------------------TCGCCAAC-ATGGGGC--G-TC---------------TATGCTAATGAT--------------GACCACTCT-GCGGATGAGTAACGCAGT---------TATATGCAATAT---GC--CCCAATAAAACAATTAAAAAATGGCA-AAGTT-------------TCTCAAAGAATATGATTCTTT-------------------------------ATTGAATA------------------------------------------------------TTGTA----------TGT---------------------------------------------------AGA---------TTGGGAAACCATAACGACTTC-CATTAA---------TAC-------------TACTCC-T--TTCTTC-CCAATAGCTCTGGTGACATGATTGTAGTGACCACCGAGTCCGAACTAACTCCAGACG | |
| dm3 | chr3R:25317911-25318167 - | TGCATCGGTCGGGGCAAGCCCAAGGCGGGCACGGCCACCATCCGTCAGCGGTAAGAGCG--------------------------------------GGCG-A------TCGCGA-TCGA--GTGTCGGCGGGTCAAT---GGGGTTAATCGGCCAGC--------------CACACTCGTT------CGTCT----------------G-TCTGGC--------------------------------------------------TATGAGTAATGCCATCATGGGCTCCAACTTGGA--GCAG---------------------------------------------------------------------C-------------------------------ATCAACTG------------------------------------------------------TTTGCT--------------------------------------------------------------------------------TAC---------------G---------------C--------CTCTCTGC----T---CTCT-GCTTGCAGCTCCGGTGACATGATCGTGGTGACCACCGAGTCGGAACTCACGCCCGATG | |
| droSim2 | 3r:24678009-24678264 - | dsi_30951 | TGCATCGGGCGCGGCAAGCCCAAGGCGGGCACGGCCACCATCCGCCAGCGGTAAGAGAG-CG-----------------------------------GGTG-A------TGGCGA-TCGG--GTGTCTGCGGGTCAAT---GGGGTTAATCGGCCAGC--------------CACACTCGTT------CGTTC----------------G-TCTGGC--------------------------------------------------TATGAGTAATGCCAT---GGGCTCCAACTTGGA--GCGC---------------------------------------------------------------------C-------------------------------ATCAACTG------------------------------------------------------ATTGCT--------------------------------------------------------------------------------TAC---------------A---------------C--------CTCTCTGC----T---CTCT-GCTTGCAGCTCCGGCGACATGATCGTGGTGACCACCGAGTCGGAACTCACGCCCGATG |
| droSec2 | scaffold_4:4176410-4176665 - | TGCATCGGGCGGGGCAAGCCCAAGGCGGGCACGGCCACCATTCGCCAGCGGTAAGAGTG-CG-----------------------------------GGTG-A------TTGCCA-TCGG--GTGTCTGCGGGTCAAT---GGGGTTAATCGGCCAGC--------------CACACTCGTT------CGTTC----------------G-TCTGGC--------------------------------------------------TATGAGTAATGCCAT---GGGCTCCAACTTGTA--GCAC---------------------------------------------------------------------C-------------------------------ATCAACTG------------------------------------------------------ATTGCT--------------------------------------------------------------------------------TAC---------------A---------------C--------CTCTCTGC----T---CTCT-GCTTGCAGCTCCGGCGACATGATCGTGGTGACCACCGAGTCGGAACTCACGCCCGATG | |
| droYak3 | 3R:21164962-21165219 + | TGCATTGGGCGTGGCAAGCCCAAGGCGGGCACTGCCACCATCCGGCAGCGGTAAG-----------------------------------------------------------A-TCGG--GTGTCTGCGGGTCAAT---GGGGTTAATCGACTGGC--------------CACATTCGTC------TG----------------C--G-TCTGGGTG----GGCCGCATGCTAATGAT--------------GAGCACGCG-GCGGATGAGTAATGCTGC---------CAATTCGGA--GTCG---------------------------------------------------------------------C-------------------------------ATCAACTA------------------------------------------------------ATCACT--------------------------------------------------------------------------------TGT---------------A---------------C-------------TGC----C---CTCT-GCTTGCAGCTCCGGTGACATGATCGTGGTGACCACCGAGTCGGAACTCACGCCCGATG | |
| droEre2 | scaffold_4820:2687260-2687528 + | TGCATTGGGCGTGGCAAGCCCAAGGCGGGCACGGCCACCATTCGGCAGCGGTAAGC------------------------------------------------------------TCGG--GTGTCTGCGGGTCAAT---GGGGTTAATCGACTGGCCTGG---------CCACATTCGTC------TG----------------C--G-TCTGGGTG----GGCCGCATGCTGATGACG-------AGCGACGACCACGCG-GCGGATGAGTAATGCTGC---------CCAGTC-AG--ACGC---------------------------------------------------------------------C-------------------------------ATCAACTA------------------------------------------------------ACCACT--------------------------------------------------------------------------------TGT---------------A---------------C-------------TGC----C---CTCT-GCTTGCAGCTCCGGCGACATGATCGTGGTGACCACCGAGTCGGAACTCACGCCCGATG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 10:02 PM