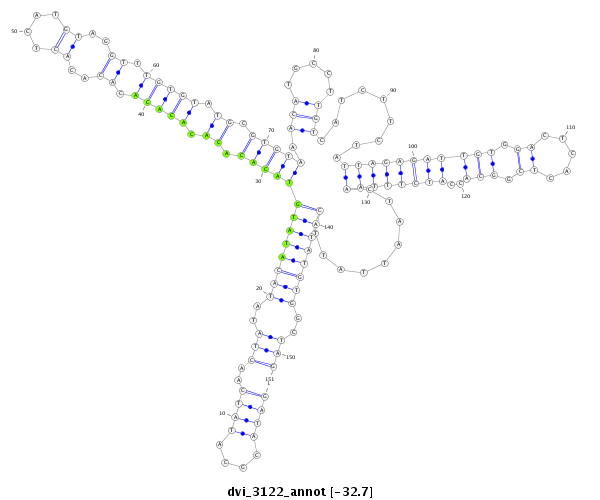
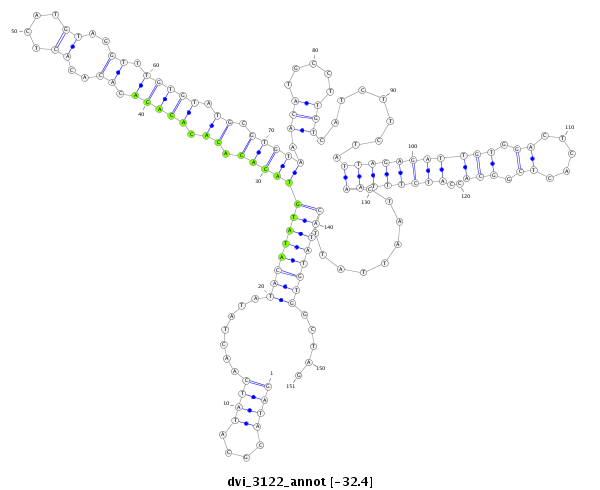
ID:dvi_3122 |
Coordinate:scaffold_12823:2215217-2215367 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
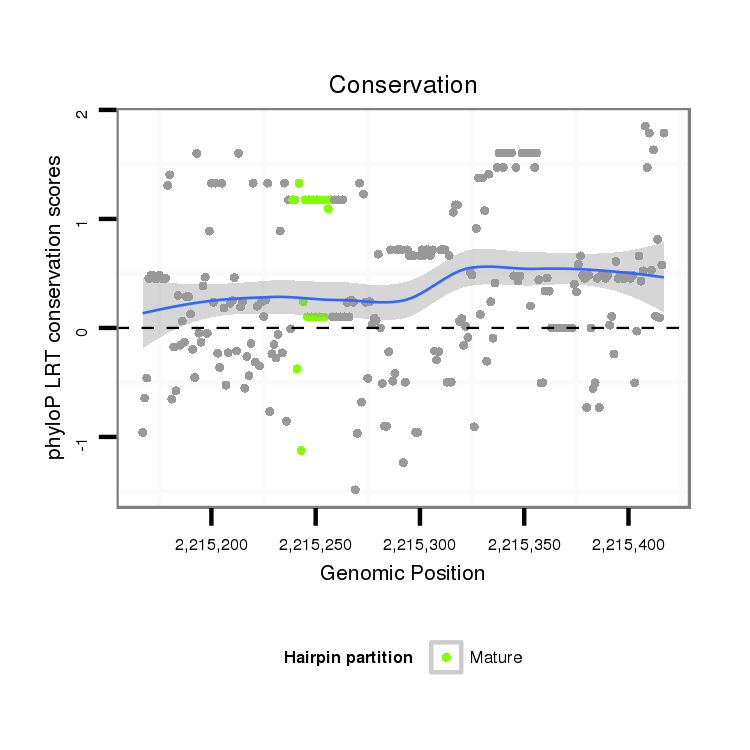
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -32.4 | -32.3 | -32.3 |
 |
 |
 |
CDS [Dvir\GJ18776-cds]; exon [dvir_GLEANR_360:2]; intron [Dvir\GJ18776-in]
| Name | Class | Family | Strand |
| (CA)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTGCTTGCAGCGCAGTAATTGTTAATTTGCAATTAATGTTCACAAATTTTGATACGCATATCAACTATATACATATGTACACACACACACACACACACTCATGTAGGTTTGTGTATGCGTGTAAACATGCCTTGTCATCTTCTATTAGAGATTGTGGACTCACTCGGCACCATCTTTAACTAATTATTCATATGTGGCTAGCTGCTCGCGCTCGTGACCGTTCTCATTCTGGATCTGGCCGACACGAACTT **************************************************((((....))))..(((..(((((((((((((.((.(((((.((..((....))..)).))))).)).))))).(((.....))).........(((((((((((.((.....)).)))..)))))))).........))))))))..)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060666 160_males_carcasses_total |
V047 embryo |
M047 female body |
SRR060664 9_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
GSM1528803 follicle cells |
M061 embryo |
SRR060663 160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
M028 head |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................GTTGACCCACTCGGCACC................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GACTCACTCGGCACCTTAC............................................................................ | 19 | 3 | 6 | 0.33 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GACCCACTCGGCACCTTCC............................................................................ | 19 | 3 | 15 | 0.27 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................ACCATCTTTACCTAACTATCC.............................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TGACCCACTCGGCACCCTCT............................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................AGGACCCACTCGGCACCTT.............................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TTAATGACTCACTCGGCAC................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................TTTTTGACCCACTCGGCACC................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GTTGTGTATGCGTGTAAAA............................................................................................................................. | 19 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................GGAAAAATGCCTCGTCATC................................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TCTAGGTTGGTGCATGCGT................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................ATATGTACACACACACACA................................................................................................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AACGAACGTCGCGTCATTAACAATTAAACGTTAATTACAAGTGTTTAAAACTATGCGTATAGTTGATATATGTATACATGTGTGTGTGTGTGTGTGTGAGTACATCCAAACACATACGCACATTTGTACGGAACAGTAGAAGATAATCTCTAACACCTGAGTGAGCCGTGGTAGAAATTGATTAATAAGTATACACCGATCGACGAGCGCGAGCACTGGCAAGAGTAAGACCTAGACCGGCTGTGCTTGAA
**************************************************((((....))))..(((..(((((((((((((.((.(((((.((..((....))..)).))))).)).))))).(((.....))).........(((((((((((.((.....)).)))..)))))))).........))))))))..)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
V116 male body |
SRR1106729 mixed whole adult body |
V047 embryo |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
M047 female body |
SRR060672 9x160_females_carcasses_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......CGTCGCGACATTAACAATTG................................................................................................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CCGTGTTAGAAATTGA...................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GAGCACTGGGAAGAGTAG....................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TGGAGCACTGGGAAGAGTA........................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................ATCGAGCCGTGGTAGATAT......................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................AGTACAGGCAAGAGTAAGT..................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................CGAGTCTTTAAAACCATGC................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................GATTTTTACGGAGCAGTAG................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................GTGTGTGTGTGTGTGAGTA..................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................AAGTACAGGCAAGAGTAAG...................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12823:2215167-2215417 + | dvi_3122 | TTGCTTGCA---GCGCAGTAATTGTTAATTTGCAATTAATGTTCACAAATTTTGATACG----------CATATCA-----------------------------ACTATATACATATGTACACACACACACACAC-ACACTCATGTAGGTTT----GTGTATGCGTGTAAACATGCCTTGTCATCTTCTATTAGAGATTGTGGACTCACTCG-----GCACCATCTTTAACTAATTATTCAT----------ATGTGGCTAGCTGCTCGCGC------TCGTGACCGTTCTCATTCTGGATCTGGCCGACACGAACTT |
| droMoj3 | scaffold_6496:1564445-1564677 + | TCGCTTGCAAGCGCGCAGTAATTGTTAATTTGCAATTAATGTTCACAAGTTTTGATACG----------CA-ATTGAGG------------------------------CAAACATGTCTATATATATATACATATTATATTTATTCATGTAT----GTG-----ATGAAAAGCTGCCCCGTCATCTTCTATTCGCGATTATA---------------GCCTCATCTTTAGCTGATTATTCATT---------TGCTC-----------GCGTGCCTTCTCTT-TTCTTTCTTGTTCTGGATCTACTCGACACGAACTT | |
| droGri2 | scaffold_15245:15863957-15864180 - | ATA------------CATATACGGCCAGCTGCAG--TAAACTTTGTGAATTTCGCTATT----------AATTTTGATATGGCGATTTTGATATAGGCAATATTTACTACAA-----TGT---------------------T-TTCAATGTATTCAAATG--ACGGTCAAAATATGCCAAGTCATCTCAAATTAGAGATTGCAGACGCAGTCA-----GCACCATCTTTAACTAATTACTCATTCGCACATATGTGCG-----------AGATCT-----------------CTTT---------CTGGACACGAACTT | |
| droWil2 | scf2_1100000004921:905278-905390 - | GCA------------CATTGATGTTTGGATTGCA--TAAAATTAACAGATTCCAAAAAG----------AGTATAGAAT------------------------------TAGATATATTCACACACACACACACAC-ACACTCATGTATGTGC----------------------------------------------------------------------------------------------------------------------GTGT-----------------GTAG---------GTGTA----GA--- | |
| droPer2 | scaffold_56:264528-264635 - | AT---------------------------TTGCATTTGATGTGTGCAAGTGTACGCACGCTCCCACTCACACATCA-----------------------------CGAACATATATGTATACACACACACACACAC-ACACACACACAAGCGT----GCAT------------------------------------------------------------------------------------------------------------------------------------------------------ATGTGTGT | |
| droAna3 | scaffold_13266:4029200-4029295 + | TGT------------CACATTCGATTGAATATCA-------------AGTG-------------------------------------------------------------------------------------------------------------------------------------------------GATGGCAT--CCACT----GGTGGGCCATCTTTAACCAATTACTCAT----------GAGTG-----------GCGTGT-----------------CTCT---------CTGGACACGGAATT | |
| droBip1 | scf7180000396427:2492989-2493039 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCATCTTTAACCAATTACTCAT----------GAGTG-----------GCGTGT-----------------CTCT---------CTGGACACGGAATT | |
| droKik1 | scf7180000302471:1027645-1027709 - | C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTCAGTAGTGGGCCATCTTTAACCAATTACTCAT----------GGGTG-----------GCGTGT-----------------CTCT---------CTGGACACGGAATT | |
| droRho1 | scf7180000780104:176658-176716 - | C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTC------GGGCCATCTTTAACCAATTACTCAG----------AGGTG-----------GCGTGG-----------------CTAT---------CTGGACACGGAATT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/16/2015 at 08:09 PM