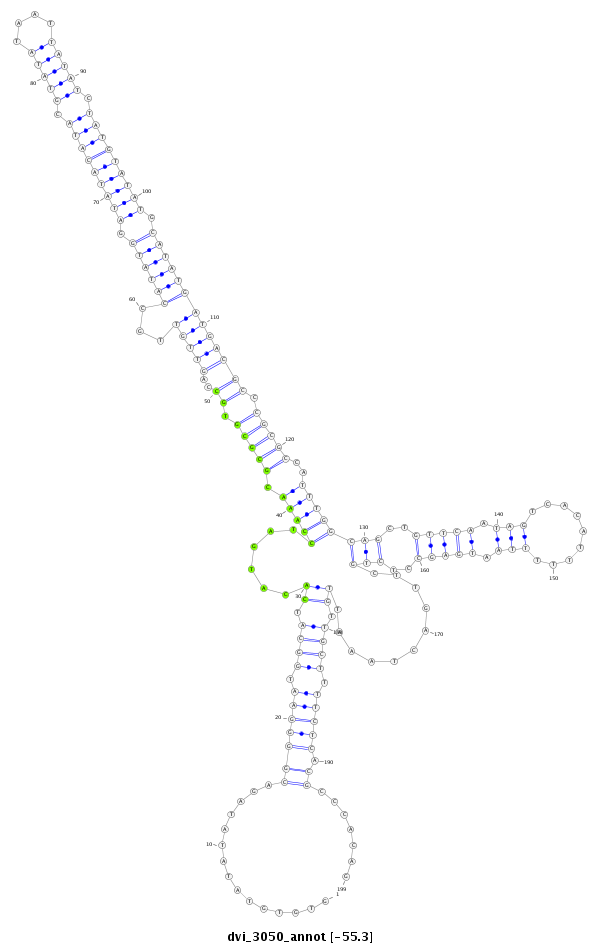
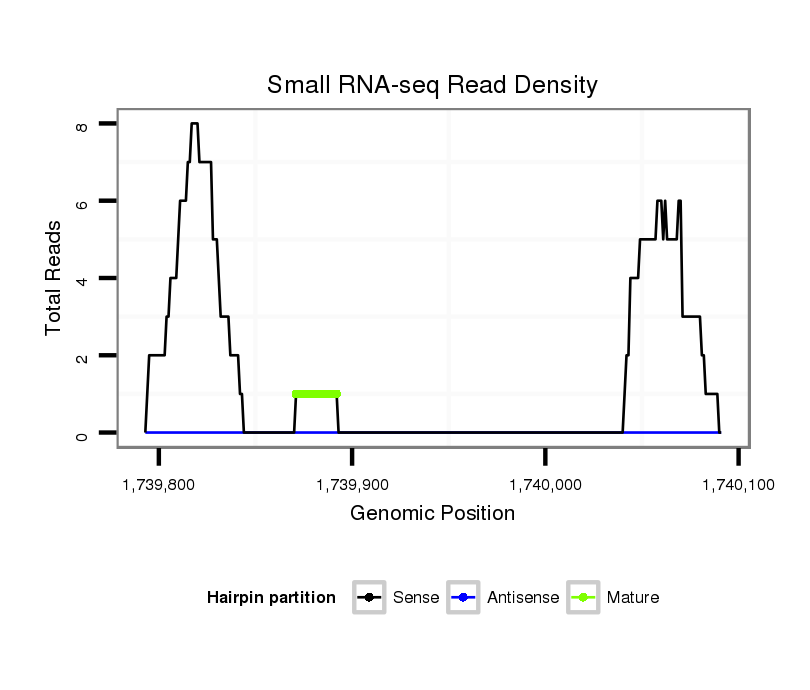
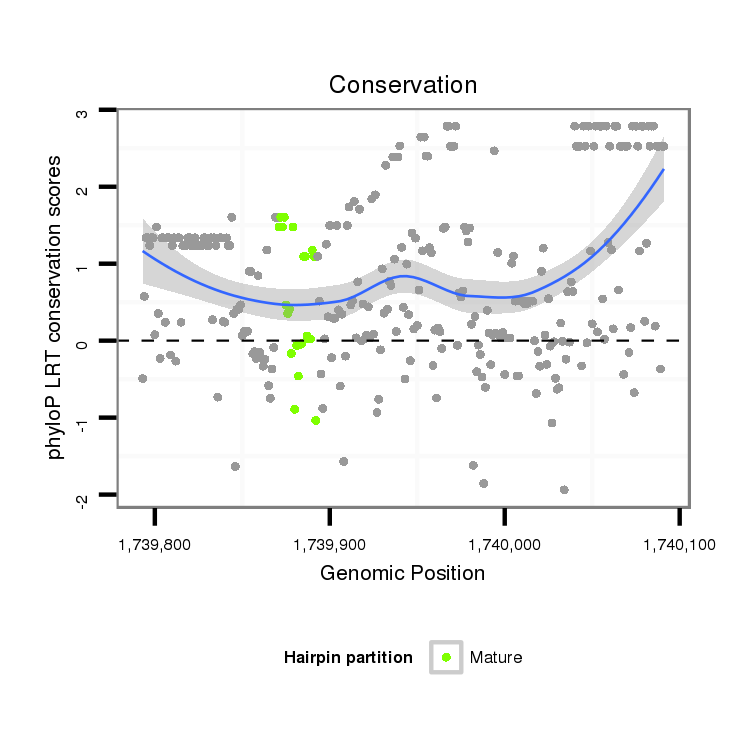
ID:dvi_3050 |
Coordinate:scaffold_12823:1739843-1740041 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -55.2 | -55.2 | -55.2 |
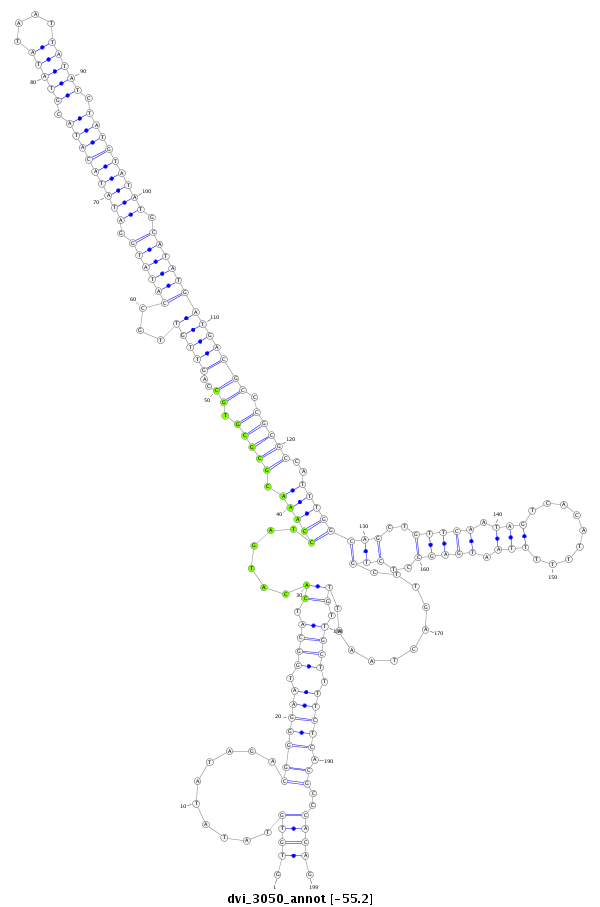 |
 |
 |
CDS [Dvir\GJ18531-cds]; exon [dvir_GLEANR_333:4]; exon [dvir_GLEANR_333:3]; CDS [Dvir\GJ18531-cds]; intron [Dvir\GJ18531-in]
| Name | Class | Family | Strand |
| (TATATG)n | Simple_repeat | Simple_repeat | + |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGAAGAACGCCTACAACGAATACCGCAAGCTGCCCATCGATACCAAACTGGTGTGTATATATAGACGGGGAATGGCATCACATGATCCAAACGCGCGTGCCAGTTGTTGCCATATGGATATACATACGTATATAATTATATCTATGTATATGCATATGATGACGCCCGCGCCATTTGGCAGCTGTTCAATAGTCACATTTTTTAATGAGCCTCTGCTTGACTAAATTGTTGCTTTTCTCACGCCCACAGGCGACCAGCGAGTATCTGGCATTTGCGGCGATATCGAAGCTGATTGCCGC **************************************************...............(((((((.((((.((......(((((.(((((.((..(((((...((((((.(((((((((.(((((....))))).))))))))).))))))))))))).)))))..)))))(((..(((((.(((.........))).)))))..)))...........)).)))).))))).)).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
M047 female body |
V047 embryo |
SRR060686 Argx9_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060685 9xArg_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060670 9_testes_total |
SRR060681 Argx9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
M027 male body |
SRR1106730 embryo_16-30h |
SRR060663 160_0-2h_embryos_total |
V053 head |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................................GACCAGCGAGTATCTGGCATTTGCGGC..................... | 27 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AATACCGCAAGCTGCCCATCG.................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GAATACCGCAAGCTGCCCATC..................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................CCAGCGAGTATCTGGCAATTGC........................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................AGACCAGCGAGTACCTGGCATTTGCGGC..................... | 28 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................GCGACCAGCGAGTATCTGG............................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................GTCGACCAGCGAGTATCTGGCATTTGC........................ | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TACAACGAATACCGCAAGCTGCCC........................................................................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................GCGATATCGAAGCTGATTGCC.. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CAACGAATACCGCAAGCTGCCC........................................................................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................TGGCATTTGCGGCGATATCGAAG........... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................ATTTGCGGCGATATCGAAGCT......... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GAAGAACGCCTACAACGA........................................................................................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................GCGAGTATCTGGCATTTGCGGC..................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................GAGTATCTGGCATTTGCGTC..................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................GCAAGCTGCCCATCGATACCAAACTGG........................................................................................................................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................CCGCAAGCTGCCCATCGATACCAAACT.......................................................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CACATGATCCAAACGCGCGTGC....................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................AGACCAGCGAGTATCTGGCATTTGCGGC..................... | 28 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..AAGAACGCCTACAACGAATACCGCAA............................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................GGCGACCAGCGAGTATCTGGCA............................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ATACCGCAAGCTGCCCATCGATACC............................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................GATGTCGAAGCTGATGGCC.. | 19 | 2 | 5 | 0.60 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................TCTGGCATTTGCGGC..................... | 15 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................AGACGGGGAATAGCACC............................................................................................................................................................................................................................ | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................ATGTCGAAGCTGATGGCC.. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................CATCTTTACCAAGCTGGTGT..................................................................................................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................ATACGAATCTAATTATAT.............................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........CATATCGAATACCGCAAGC............................................................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACTTCTTGCGGATGTTGCTTATGGCGTTCGACGGGTAGCTATGGTTTGACCACACATATATATCTGCCCCTTACCGTAGTGTACTAGGTTTGCGCGCACGGTCAACAACGGTATACCTATATGTATGCATATATTAATATAGATACATATACGTATACTACTGCGGGCGCGGTAAACCGTCGACAAGTTATCAGTGTAAAAAATTACTCGGAGACGAACTGATTTAACAACGAAAAGAGTGCGGGTGTCCGCTGGTCGCTCATAGACCGTAAACGCCGCTATAGCTTCGACTAACGGCG
**************************************************...............(((((((.((((.((......(((((.(((((.((..(((((...((((((.(((((((((.(((((....))))).))))))))).))))))))))))).)))))..)))))(((..(((((.(((.........))).)))))..)))...........)).)))).))))).)).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060663 160_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
V053 head |
V116 male body |
SRR060665 9_females_carcasses_total |
V047 embryo |
M061 embryo |
SRR060660 Argentina_ovaries_total |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................CCGCTTGTCGCTCATACACCTTAA........................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................CGCTTGTCGCTCATACACC............................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................GCGGGGGTGTCCGCTGATCG......................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................GGACCGTATACGCAGCTATAG............... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................TCGCCAGGTTATCAGTTTAAA................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................AGGGGGTGTCCGCTGGTAG......................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................GGAGGTGTCCGCTGATCGC........................................ | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................GTCATCGCCGGTATAGCTTC........... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................CTGGCTTTCGACGGGTTGCT................................................................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................TGCAGATTACCGTAGTGTA........................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12823:1739793-1740091 + | dvi_3050 | TGAAGAACGCCTACAACGAATACCGCAAGCTGCCCATCGATACCAAACTGGTGTGTATATATAG---------ACGGGGAAT---------------GGCATCACATGATCCAAACGCGCGTGCCAGTTGTTGCCATATG----GATA--TACA--TACGT-----------AT----------ATAATTATA------TCT--ATGTATATGC-------------------------A-----------TA-----------TGATGACGCC-------CGCGCCA----TTTGGCAGCTGTTCAATAGTCACA----T-------------TTTTTAA---------------------TGAGCCTC----------------------------TGCTTGACTAAATTGTTGCTTT-----------TCTCACGCCCACAGGCGACCAGCGAGTATCTGGCATTTGCGGCGATATCGAAGCTGATTGCCGC |
| droMoj3 | scaffold_6496:1033587-1033884 + | TGAAGAACGCATATAATGAATATCGCAAGCTGCCCATCGACACGAAATTGGTGTGTA-----------------------------------------ACATCACATGATCCA-------------GCTGTTGCCAACTA----GATA--TATAGCTACAGATGTCGTATAT------------------------------------AGCGCGTGCGATCGCATATGTATATATATGTATATAGTATAGCTATATATG-------------------------CCTA----TTTGGCAGCTGTTCAATTTATTTT---------T---------------------------ATTTTCAACTAAATT-CAACATTCCTCTCTCTCTCTCTCGCC-----------------------------GT------CTCCCTCTATTAGGCAACGAGTGAATATTTGGCATTTGCGGCGATGTCGAAGCTGATTGCAGC | |
| droGri2 | scaffold_15245:16298499-16298843 - | TGAAGAACGCCTACAACGAGTACCGCAAGCTGCCCATCGATACCAAACTGGTGTGTATATATAA-----TCATGTGAATGACATTAATGAGCTGTTTAACATCACATGACCGAAACGCGGCTGGCATACGTTCCCATATG----CATA--TA-----------------------TCATTATATATATATATATCTATAT----AGATATATGT-------------------------A-----------TA-----------TGATGACGCCAGT----TGTGTTT----TTTGGCAGCTGTTCAACAGCCAAAAATTT----AATGGC-CTCGCTCT------------------------C---------------------------GATATATATTTGACTAA-ACTTTGCCCACTTTCTGCTTATCCCATGACAACAGGCGACCAGCGAATATTTGGCATTTGCTGCCATGTCGAAACTGATTGCCGC | |
| droWil2 | scf2_1100000004954:5270386-5270663 - | TGAAGAATGCTTACAATGATTACCGCAAGCTGCCCATCGATACCAAGCTGGTGAGTGGGTGTAGCCAA------------AA---------------CGAATCACATGACCAAC--GCGCGTGGC---------------------TA--TATATA--------------------------------TGATA------TTTGCATATATCCGG-------------------------C-----------CGTATCTGGTTGGCGGTGACGTCGGTGTGGCGTGGGA----TTTGGCACTTGTTCAACATTATT-----T-------------CTCTAAC---------------------CGAATTTCAAAT---------------TTTGTT----------------------TCCC-----------TCGCTCTCTTTAGGCTACTACAGAATATTTGGCCTTTGCCGCTATATCCAAATTGATAGCTGC | |
| dp5 | 3:14686174-14686492 - | TGAAGAATGCCTACAACGAATACCGCAAGCTGCCCATCGATACGAAACTGGTGGGTATACA------AATCA-GTGGAGAAC---------------AGCATCACATGACCAGGTTGCTCGTGCC----GTTGCCATCCAATATAATA--TATACATGCAGACATAGTATAGGC----------ACATCCATA------TCTGTATATATCCGG-------------------------C-----------TGTGGCTGCTCTTTGGTGACGTC-------TGAGCGA----TTTGGCAGCTGTTCAAGAAAATT-----T--------------------------CTTTTCGCT------CGCATTTCTAA-------------------------------------CCGTTGATCGC-----------TTTCCATCCACAGGCCACCACCGAGTACTTGGCCTTTGCAGCCGTCTCCAAGCTGATTGCGGC | |
| droPer2 | scaffold_4:2929355-2929673 + | TGAAGAATGCCTACAACGAATACCGCAAGCTGCCCATCGATACGAAACTGGTGTGTATACA------AATCA-GTGGAGAAC---------------AGCATCACATGACCAGGTTGCTCGTGCC----GTTGCCATCCGATATAATA--TATACATGCAGATATAGTATAGGC----------ACATCCATA------TCTGTATATATCCGG-------------------------C-----------TGCGGCTGCTCTTTGGTGACGTC-------TGAGCGA----TTTGGCAGCTGTTCAAGAAAATT-----T--------------------------CTTTTCGCT------CGCATTTCTAA-------------------------------------CCGTTGATCGC-----------TTTCCATCCACAGGCCACCACCGAGTACTTGGCCTTTGCGGCCGTCTCCAAGCTGATTGCGGC | |
| droAna3 | scaffold_13266:6122752-6122880 - | A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAATGACGTC-------CAAAGAATA--TTTGGCAGCTATCCAGAAA----------------TCGC-CTCATTAATACATT-----------------------------------------------AT-------------------TTTTTT-----------GCTTCCACCCACAGGCCACCACCGAATACCTGGGCTTCGCCGCCGTCTCCAAGCTGATCGCGGC | |
| droBip1 | scf7180000395839:279892-280027 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TA-----------TAATGACGTC-------CAAAGAATATTTTTGGCAGTTGTTCAGAAAATATC---------GAACGC-CTCACTGACATATA-----------------------------------------------TT----------------------TTT-----------GCTTCCACCCACAGGCCACCACCGAATACCTGGGCTTCGCAGCCGTCTCCAAGCTGATTGCGGC | |
| droKik1 | scf7180000302476:2333010-2333164 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A------TCCGAATATATCCGG-------------------------------------TG-----------TGATGACGTC-------TCTGCGA----TTTGGCAGCCATTCAATAAATACG---------AAACGCCCCCACTAATATATGTATT---------------------------------------CTCGTT-----------------------------CT------CTCCATCCCACAGGCCACCACCGAGTACCTGGCGTTTGCGGCCATCTCCAAGCTGATCGCAGC | |
| droFic1 | scf7180000453851:1205053-1205241 + | TG--------------------------------------------------------------------------------------------------------------------------------CTATTATTTT----GAT---CACA--TCTGT-----------ATTCCACCATATATATGCATA------ACCGAATTTATCTGG-------------------------T-----------TG-----------TGATGACGTA-------CCTGCGA----TTTGGCAGCCATTCAGCACATATC---------GTTCGC-CTCACTAATATATG-----------------------------------------------TT-------------------------------------TTCTCTCCCACAGGCCACCACCGAGTATTTGGCATTCGCGGCCGTTTCCAAGCTGATCGCGGC | |
| droEle1 | scf7180000491201:892422-892626 - | AT------GA-----------------------------------------TTCAGA-----------------------------------------AAATCACACTCTCTC-------------ATTGCTATTATTTT----GATGAACACA--TCTGT-----------AC----------ACAACCATA------ACCGAATATATCCGG-------------------------C-----------TG-----------TGATGACGTC-------TCTGCGA----TTTGGCAGCCATTCCGT-----------------TTTGC-CTTACTAATATATG-----------------------------------------------TA----------------------TTCC-----------TTTTTACACACAGGCAACTACCGAGTACCTGGCCTTCGCGGCCGTGTCCAAGCTGATCGCGGC | |
| droRho1 | scf7180000773652:169448-169611 - | T------------------------------------------------------------------------------------------------------------------------------------------------------ACA--TCTGT-----------AC----------ACAACCATA------ACCGAATATATCCGG-------------------------C-----------TG-----------TAATGACGTC-------TCTGCGA----TTTGGCACCCACTCAGAACATTCC---------CTTCGG-CTCGCTAATATCCG-----------------------------------------------TT-------------------------------------CCCTCTTCCACAGGCCACCACTGAGTACTTGGCCTTCGCGGCCGTTTCCAAGCTGATCGCGGC | |
| droBia1 | scf7180000301506:1656514-1656676 + | CG--------------------------------------------------------------------------------------------------------------------------------------------------------------T-----------AC----------ACAAGCATA------TCCAAATATATCCGG-------------------------T-----------TG-----------TAATGACGTC-------GCAACGA----TTTGGCAGCCATTCAATACGTATT----GCTCTGTACCC-CTTGCTAATATGTG-----------------------------------------------TT-------------------------------------TCCTCCTCCACAGGCCACCACCGAGTACTTGGCCTTCGCGGCCATCTCCAAGCTGATCGCGGC | |
| droTak1 | scf7180000411009:9307-9445 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATATCTGG-------------------------T-----------TG-----------TGATAACGTC-------GCTACGA----TTTGGCAGCCATTCAATACATTCT---------GTTCGT-CTCGCTAATATGTG-----------------------------------------------TT-------------------------------------CTCTCTCCCACAGGCCACCACCGAGTACTTGGCCTTCGCGGCTGTCTCCAAACTGATCGCGGC | |
| droEug1 | scf7180000409183:717963-718125 - | A-------------------------------------------------------------------------------------------------------------------------------------------------------CT--TCTGT-----------AC----------ACAACCATA------TCCGAATTTATCCGG-------------------------C-----------TG-----------TGATGACGTC-------TCTGCGA----ATTGGCAGCCATTCAGAATATTCT---------GTTCGC-CTCATTAATATGTT-----------------------------------------------TT-------------------------------------CCTTCTCCCACAGGCCACTACCGAGTACTTAGCCTTCGCGGCTGTCTCCAAGCTGATCGCGGC | |
| dm3 | chr2R:5075721-5075879 - | A-------------------------------------------------------------------------------------------------------------------------------------------------------CA--TCTGT-------------------------ATCTGTA------TCCGAATATATCTGG-------------------------T-----------TG-----------TGATGACGTC-------TCCGCCA----TTTGGCAGCCATTCAGCTCATTCT---------GTTCGC-CTCACTAATGTGTG-----------------------------------------------CT-------------------------------------CCCTCACCCACAGGCCACCACCGAGTACTTGGCCTTCGCGGCTGTCTCCAAGCTGATCGCAGC | |
| droSim2 | 2r:5879016-5879154 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATATCTGG-------------------------T-----------TG-----------TGATGACGTC-------TCCGCCA----TTTGGCAACCATTCAGCTCATTTT---------GTTCGC-CTCACTAATATGAG-----------------------------------------------TT-------------------------------------CTATCACCCACAGGCAACCACCGAGTACTTGGCCTTCGCGGCTGTCTCCAAGCTGATCGCAGC | |
| droSec2 | scaffold_1:2708105-2708243 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATATCTGG-------------------------T-----------TG-----------TGATGACGTC-------TTCGCCA----TTTGGCAGCAATTTAGCTCATTTT---------GTTCGC-CTCACTAATATGCG-----------------------------------------------TT-------------------------------------CCCTCACCCACAGGCAACCACCGAGTACTTGGCCTTCGCGGCTGTCTCCAAGCTGATCGCAGC | |
| droYak3 | 2L:17731092-17731248 - | A---------------------------------------------------------------------------------------------------------------------------------------------------------------T-----------AC----------ACATCCAAA------TCCGAATATATCTGG-------------------------T-----------TA-----------TGATGACGTC-------TCAGCCA----TTTGGCAGCCATTCAGCGCATTGT---------GTTCGC-CTCACTAATATGTG-----------------------------------------------TT-------------------------------------TCCTCACCTACAGGCCACCACCGAGTACTTGGCCTTCGCGGCTGTCTCCAAGCTGATCGCGGC | |
| droEre2 | scaffold_4929:17580110-17580266 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC----------ACAACCACA------TCCGAATATATCTGG-------------------------T-----------TA-----------TGATGACGTC-------TCAGCCA----TTTGGCACCCATTCAGCGCACTCT----G----TTTCGC-CTCACTAATATGTG-----------------------------------------------TT-------------------------------------CCCTCTCCCACAGGCCACCACCGAGTACTTGGCCTTCGCGGCTGTCTCCAAGCTGATCGCGGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 12:05 PM