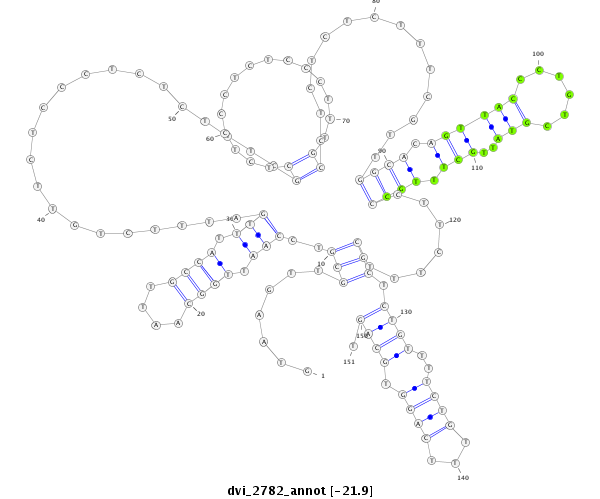
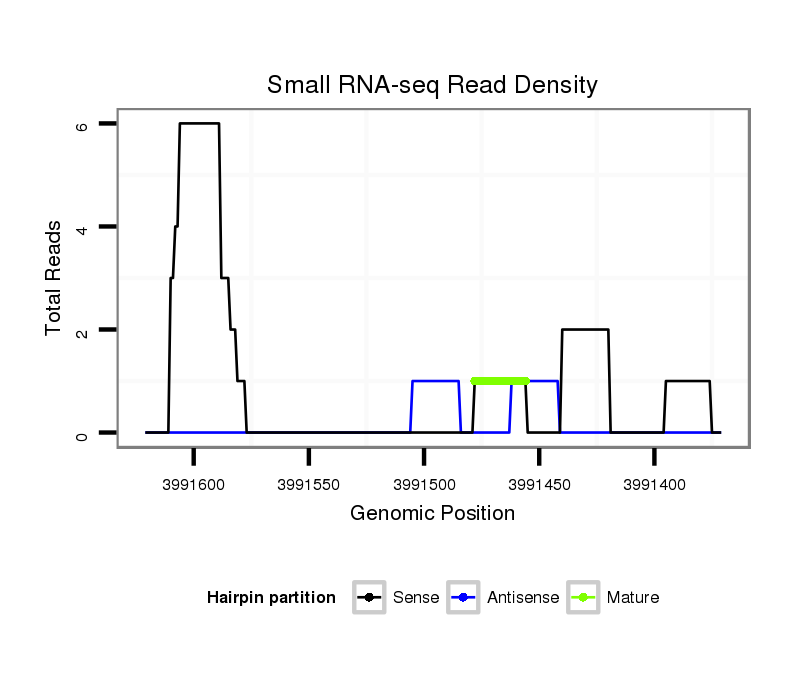
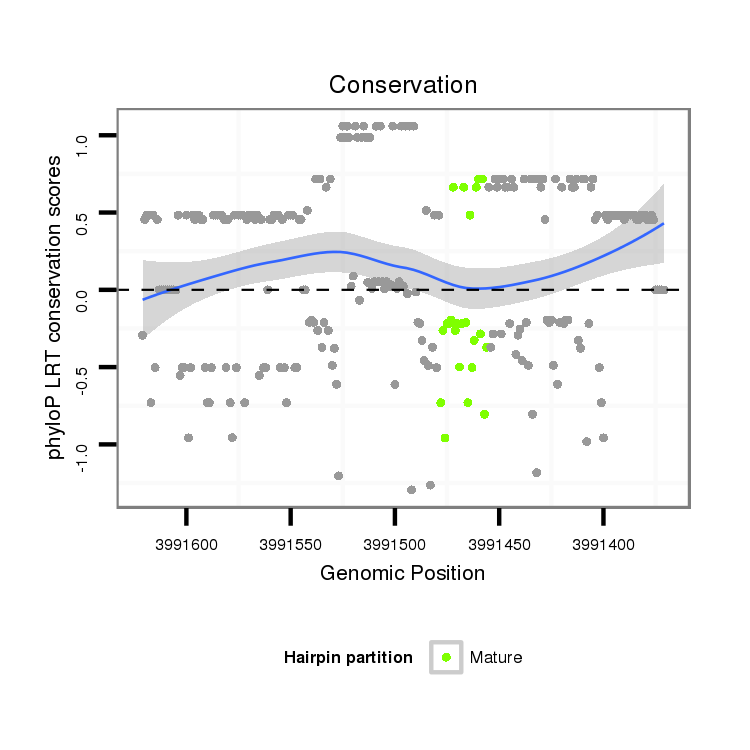
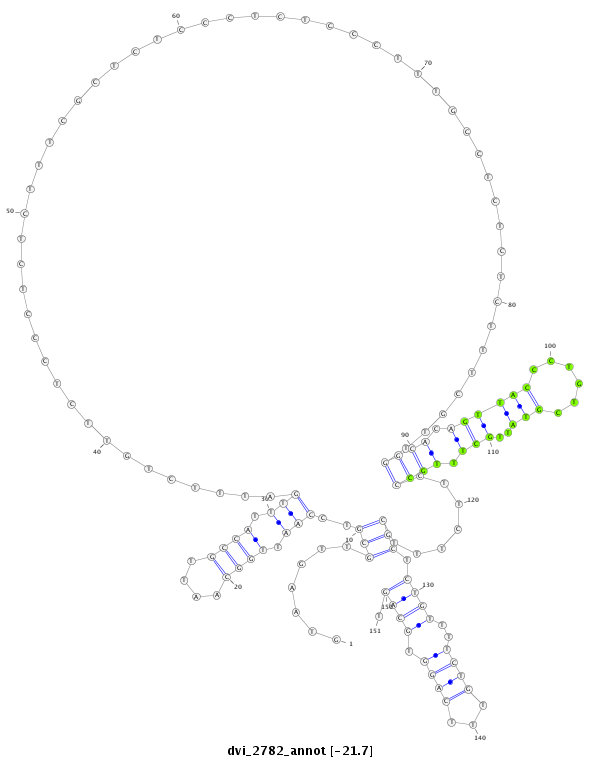
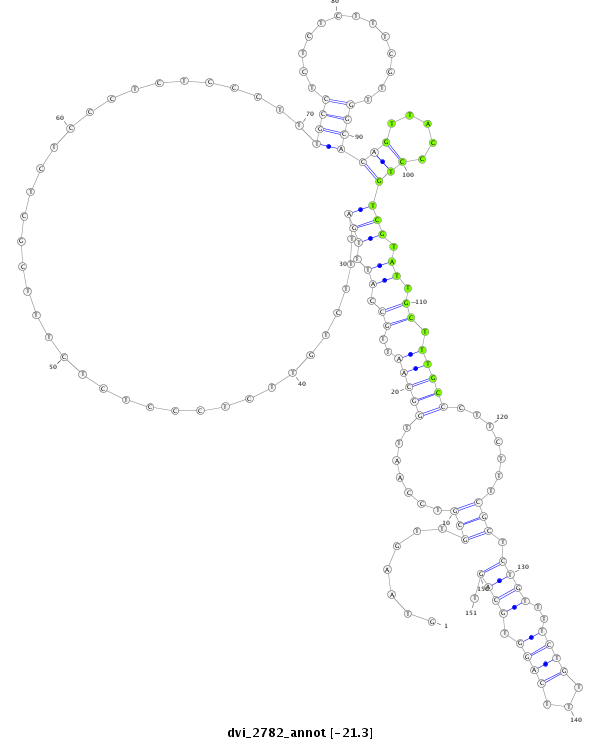
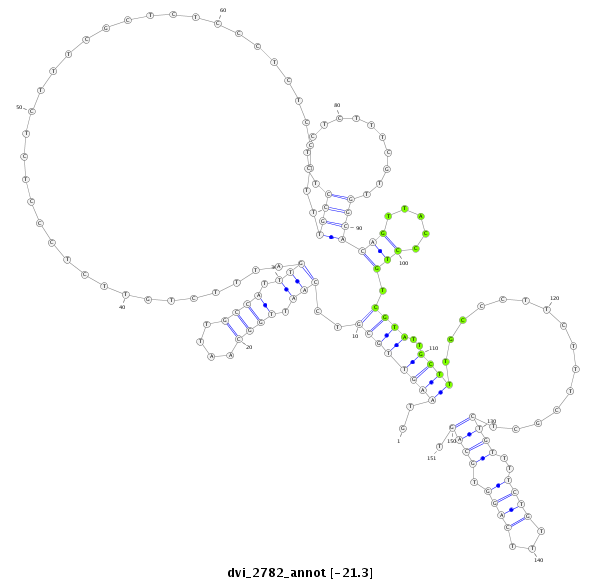
ID:dvi_2782 |
Coordinate:scaffold_12822:3991421-3991571 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -21.7 | -21.3 | -21.3 |
 |
 |
 |
exon [dvir_GLEANR_14111:1]; CDS [Dvir\GJ14158-cds]; intron [Dvir\GJ14158-in]
| Name | Class | Family | Strand |
| G-rich | Low_complexity | Low_complexity | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCAAAAGGAGCCCAATTTTGCTGCAGTTGCGCCGAAAAACGGACTCAAAAGTAAGTTGCGTCCAATTGGCAATTGCCATTTGATTTCTGTTCTCCCTCTCTTTCGCTCTCCCTCTCCCTTTGCCTCTCTCTTTCGTTGGCACAGTTACCCTGTCGTATTGCTTTGCCCTTCTTTCGCTCTGTTTTCTGTTTCAGGTGCAGTGCCCAGCTGATTGCCAGATTGTCAATAATTCAATAACCTTTGCACAGAAT **************************************************.......(((..(((.((((....)))).)))......................((...............))..............((((.((((((......)))..))).)))).......))).((((..((((...)))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
M028 head |
V047 embryo |
V116 male body |
SRR060664 9_males_carcasses_total |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........CCAATTTTGCTGCAGTTGCGCC.......................................................................................................................................................................................................................... | 22 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................TTTTCTGTTTCAGGTGCAGTG................................................. | 21 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GTTACCCTGTCGTATTGCTTTGC..................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GAAGCGTCAAATTGGCAATTGC............................................................................................................................................................................... | 22 | 3 | 3 | 1.00 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ...............TTTTGCTGCAGTTGCGCCGAAA...................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TAATTCAATAACCTTTGCAC..... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................GCAGATGCGCCGAAAAACGGA................................................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGTCGCATTGCTTTGCCCTTCTT.............................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............TTTTGCTGCAGTTGCGCCGAAAAACGGAC............................................................................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............AATTTTGCTGCAGTTGCGCCGAAAAAC................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........ACAATTTTGCTGCAGTTGCGCC.......................................................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................AGCGTCAAATTGGCAATTGC............................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CGTATAGCTTTGCCCCTC................................................................................ | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................AAGCGTCAAATTGGCAATTGC............................................................................................................................................................................... | 21 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CCTTCGATCGCTCTGTTT................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGTTTTCCTCGGGTTAAAACGACGTCAACGCGGCTTTTTGCCTGAGTTTTCATTCAACGCAGGTTAACCGTTAACGGTAAACTAAAGACAAGAGGGAGAGAAAGCGAGAGGGAGAGGGAAACGGAGAGAGAAAGCAACCGTGTCAATGGGACAGCATAACGAAACGGGAAGAAAGCGAGACAAAAGACAAAGTCCACGTCACGGGTCGACTAACGGTCTAACAGTTATTAAGTTATTGGAAACGTGTCTTA
**************************************************.......(((..(((.((((....)))).)))......................((...............))..............((((.((((((......)))..))).)))).......))).((((..((((...)))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060664 9_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
GSM1528803 follicle cells |
V116 male body |
SRR060659 Argentina_testes_total |
M027 male body |
M047 female body |
SRR060669 160x9_females_carcasses_total |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................AGAACCAAACGGGAAGAA.............................................................................. | 18 | 2 | 5 | 9.80 | 49 | 20 | 16 | 0 | 0 | 5 | 4 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GGAAACGGAGAGAGAAAGCAA.................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGAAACGGGAAGAAAGCGAGA....................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTTGCCTGAGTTGTC........................................................................................................................................................................................................ | 15 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CGGGTTAAGACGTCGTCA................................................................................................................................................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................AGAACGAAACGGGAAGAAG............................................................................. | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GCAGTTTAGTCGTTAACGGT............................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TAAGAGGGATAGAAAGCGA................................................................................................................................................ | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AAACGGAGCCGGAAAGCAAC................................................................................................................. | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................CGAGACTGGAAGGAAGCGA......................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................CACTAAAGACAAGAGGTAC......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................GAGGGAGAGGACAACGGGGAG........................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12822:3991371-3991621 - | dvi_2782 | GCAAAAGGAGCCCAATTTTGCTGCAGTTGCGCCGAAAAACGGACTCAAAAGTAAGTTGCGTCCAATTGGCAATTGCCATTTGATTT---------------------------------------CTG------------------------------------------TTCTCCCTCTCTTTCGCTCTCCCTCTCCCTTTGCCTCTCTCTTTCGTT----------------------GGCACAGTTACCCT-----GTCGTATTGCTTTGCCCTTCTTTCGCT--CTG----TTTTCTGTTTCAGGTGCAGTGCCC------------AGCTGATTGCCAGATTGTCAATAATTCAATAACCTTTGCACAGAAT |
| droMoj3 | scaffold_6540:17270088-17270437 - | GCAACAAG---------TAATTCTAGTTGCAAAAAAAAACAGCGTTAAACGTAAGTAGTA-CCAATCGAAAATCACC---TGATATTTAACAAATCGAATGTGCAACAAAGTTGAAAAAAACATTTTGTTTAATAAATTGTTTCATACTAAATGAAACCAAAGAAGCTTTATCTGTCTCTCTTTCGCTCCCTTTTTTTTGCTGTTTATGTATTTTT--AAAAATTTTAATTAAAATTATTGCTATATAAATCCAAAGAAAGTGTCTCTCTCTCTCTCTCTCTCGTT--CAG----C-TTCTCTGTCAGGTGCACTGTCCTATAAATTGGCAAGCTTATTATCAGACGCTCAATAATTCAATAACCTTTGCAC----- | |
| droGri2 | scaffold_14853:5624883-5625019 + | T------------------------------------------------------------------------------TCAGTTT---------------------------------------TCG------------------------------------------TTGGCGCTCTCATTCGCTCTCTCTCTCTCTCTCTCTCTCTGTGCCGCT----------------------CTT----TCTCTCT-----GTCGC---GCTCTCTCTCTCTCTCGCTGTCATTTTGTCTCGTCTCTCA-ACAGAGCGCTT------------AGCTGCTTTCCA---------------------------------- | |
| droPer2 | scaffold_0:366488-366524 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCTCTCTCCCTCTCTCTCTCTCTCTCTCTCTCTCTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/16/2015 at 09:26 PM