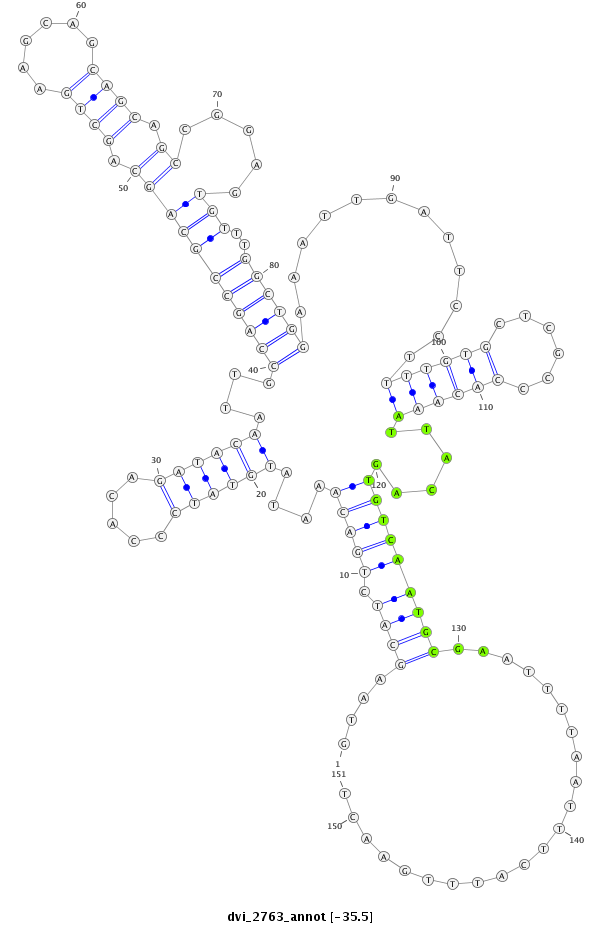
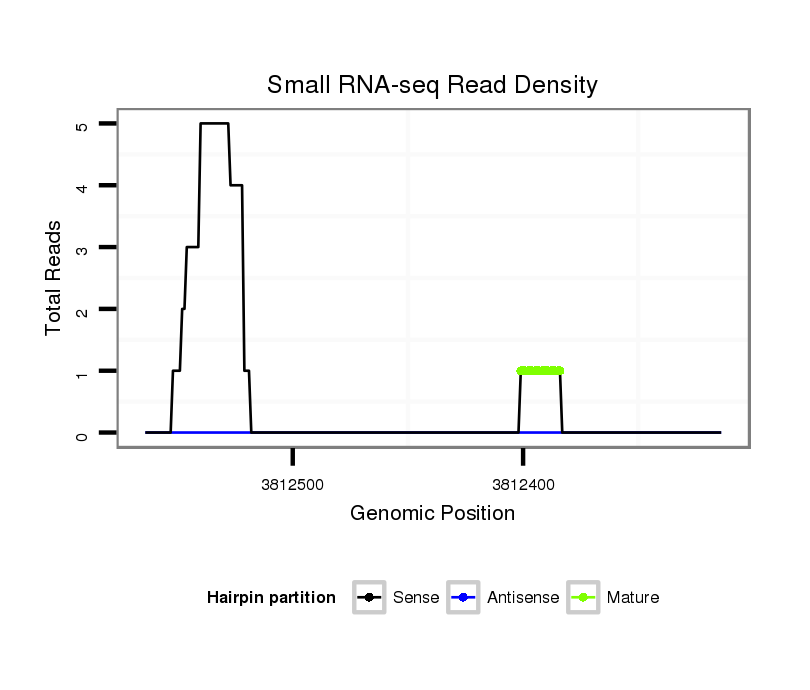
| droVir3 |
scaffold_12822:3812314-3812564 - |
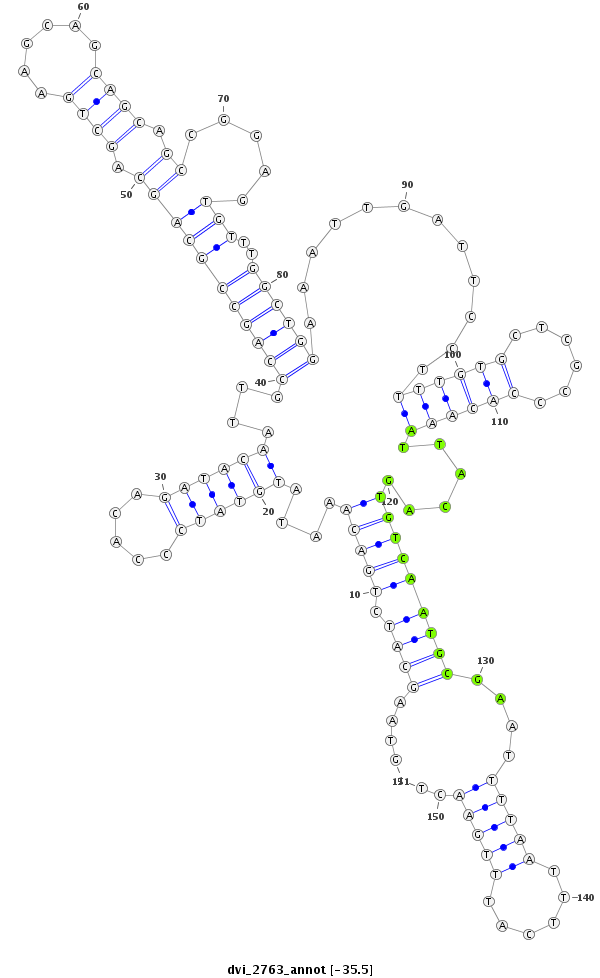
dvi_2763 |
GTATAA-----------A-G----CA-------GCAGC-----AGCAGCA-GC-GAAATGGATTTATCGAGGCGGTTGCGGTAAGCATCTGACAAATATGTATCC-------------------------------------------------------------CACAGA----T------------------------ACAATTGCCA---------------GCCGCAGCAGCT------------------------GAAGCAGC------AGCAGC---------------------CG-------GAGTGTT-T-----GGCTGGAAATTGATTCCTTTTGTGCTCGCCCACAAATTACAGTGTCAATGCGAATTTTAATTTCATTTGAACTACATGTGCAAACACTTGACTTGG----C--TCCCAACT-GGCA--GC-CT--G--GCAGCCTG---------------------------------------A-------- |
| droMoj3 |
scaffold_6540:17121221-17121495 - |
|
CTATAT-----------A-G-------------ATAGCA-----CCAGCG-GC-GAAATGGATTTATCGAGGCGGTTGCGGTAAGCATCTGACAAATATGTATCT-------------------------------------------------------------CATAGA----T------------------------ACAATTGCCA---------------GCCGCAGCATCGGCAGCAGCGGCAACAGCGGCAGCAGCAGCG------GCAGCAGT---------------------TG-----GATTGTGTT-T-----GGCACGAAATTGATTCCTTTTGTGCTCGCCCACAAATTACAGTGTCAATGCGAATTTTAATTTCATTTGAACTACATGTGCAAACACTTGACTTGG----C--TCCTGACT-GGCT--GC-CA--G--GCATCCTG---------------------------------------C-------- |
| droGri2 |
scaffold_14906:1405192-1405413 - |
|
ATC-----------------------------------------GAAGCA-GC-GAAATGGATTTATCGAGGCGGTTGCGGTAAGCATCTGACAAATATGTATCC-------------------------------------------------------------CACAGA----T------------------------ACAATTGCCA---------------GCCGCAGCAGCAGCCGCAGTAGCAT--------------------------------------------------------CAGC-TCGTATT-T-----GGCGTAAAATTGATT-----------------CAAATTACAGTGTCAATGCGTATTTTAATTTCATTTGAACTACATGTGCAAACACTTGTCTTGG----C--TCCTGACT-GGCT--GG-CT--G--ACTGGCTG---------------------------------------G-------- |
| droWil2 |
scf2_1100000004902:9045940-9046207 - |
|
GAG--C-----------T-GCTTCTGCTGCT-G----CAATCTTCAAGTG-ACAAAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCTGACAAACAA-CATCCATGTACA-----------TATATCCCCCTTCTTCCTTCCATCTATC--------GAAATGGCA----------------------------------------------------------------------------------------------------------------------------------AACTGT-----GT-TCGTGTT-T-----GGCATGAAATTGATTCCTTTTGTG-TGGCCCGCAAATTACTGTGTCAATGGGAATTTTAATTTCATTTGAACTACATGGGCAAACACTTGATATGTCG--C--TCCTGGCT-GGTT--GCCAT-----CA-TCCTT---------C-----------------------------A-------- |
| dp5 |
2:10159523-10159870 + |
dps_454 |
TTGTGC-----------T-A----TCA---TCAACAGTA----AGGAGCC-GC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCAGACAAACATGAAA-CATGTATCTTCTGGATGGGAATAC------------------TCTAC-----------------CAGA----TGCGGCACGGCACAGCCAGATAAATACAATTGTAACCAGTGACTGTGGCAGTGGTAGTGGCA---------------GCGGCAGTAGCAGTG------GCAG---TGGCAGATACAGTGGCAAGCTGT-----GTGTCGTGTT-T-----GGCACGAAATTGATTCCTTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTTCATTTGAACTACATGCGCAAACACTTGACATGG----C--TCCTGGCT-GGCT--GG-AT--G--GTCTCC-----------CA----------------------------G-------- |
| droPer2 |
scaffold_0:10322028-10322363 - |
|
TTGTGC-----------T-A----TCA---TCAACAGTA----AGGAGCC-GC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCAGACAAACATGAAA-CATGTATCTTTTGGATGGGAATAC------------------TCTAC-----------------CAGA----TGCGGCACGGCACTGCCAGATTAATACAATTGTAACCAGTGACTGTGGCAGTGGTAGTGGCA---------------GCGGC------------------AG---TGGCAGATACAGTGGCAAGCTGT-----GTGTCGTGTT-T-----GGCACGAAATTGATTCCTTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTTCATTTGAACTACATGCGCAAACACTTGACATGG----C--TCCTGGCT-GGCT--GG-AT--G--GTCTCC-----------CA----------------------------G-------- |
| droAna3 |
scaffold_13340:17169752-17170003 - |
|
TTGTCC-----------T-G----CCA---CAGACAGCA----AGGAGCC-GC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATCC-------------------------------------------ATC--------CAATCGCGACTGA----TGTAG------------------------CTGCCA---------------TCCG--------------------------------------GGAG---------T---------------------TGAGGAGT-TCGTGTT-T-----GGCATGAAATTGATTCCTTTTGTG-TTGCCAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGCGCAAATACTTGACACGG----C--TCCCGACT-GGAT--GC-TT----TGC-TCCTG------------------------------------------------ |
| droBip1 |
scf7180000396424:671945-672217 - |
|
TTGTCC-----------T-G----CCA---CCGACACCA----AGGAGCC-GC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATCC---------------------AT------------------CCATC--------CAATCGCGACAGA----TGTAG------------------------CTGCCA---------------TCCG----------------------------AGTA------GGAGTGGC---------------------------TG--GAGT-TCGTGTT-T-----GGCATGAAATTGATTCCTTTTGTG-TTGCCAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGCGCAAACACTTGACACGG----C--TCCCGACT-GGCT--GC-TT----TGC-TCCTGCTGACGTCCCT----------------------------A-------- |
| droKik1 |
scf7180000302256:130753-130998 - |
|
AGACAGAGCCAGTGAGAGCGA---GTG---GCGACAACA----AGGAGCC-GCAAAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATCC-------------------------------------------ATT--------CGAACGCGACGGA----TGTGG-------------------------------------------------------------GGC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TAGCCAACAAATTACTGTGTCAATGGGAATTTTAATTCCGTTTGAACTACATGCGCAAACACTTGACACGG----C--TCCCGACT-GGCT--GC-TTCTGCTGC-TCCT------------------------------------------------- |
| droFic1 |
scf7180000453581:252454-252713 + |
|
TTGTGC-----------G-GGG--TCC---T---CAGCA----AGGAGCCTGC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATCCATCCATCC-----------A----------------GCCATCTATCCAGCCACCCGAACGCGACAGA----TGTAG-------------------------------------------------------------TAG------------------C---------------------------------------------------T----CTGCAGTCCCTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGCGCAAACACTTGACACGGGG--C--TCCCGGCT-GGCA--GC-TC--C-AGC-TCCTG---------------------------------------C-------- |
| droEle1 |
scf7180000491280:3568182-3568413 + |
|
GTGAGA-----------G-G----AC-------TCAGCA----AGGAGCC-GC-AAAATGGATTTATCGAGGCGGCTGTGGTAAGCATCCGACAAACATGTATC-------------------TGTAT------------------CCTTC--------AGAACGCGACTGACTGGTGTGG------------------------CC-C---CA---G------------------------CAG----------------------------------------------------------------------------TGCAATCGTTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGGATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGTTTC--TCC-------------------------TCCTG---------------------------------------CTA------ |
| droRho1 |
scf7180000779501:82425-82669 - |
|
TTGTGC-----------T-G----TCC---T---CAGCA----AGGAGCC-GC-AAAATGGATTTATCGAGGCGGCTGTGGTAAGCATCCGACAAACATGTATCCATC---------------CATAT------------------CCATC--------CGAACGCGACAGA----TGTAG------------------------C------CAGTGG------------------------CAG----------------------------------------------------------------------TG----TGCAATCGTTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGG--C--TCCCAGTT-GGCT--GG-------TGC-TCCTG------------------------------------------------ |
| droBia1 |
scf7180000302075:2418959-2419210 + |
|
GTGTCC-----------C-G----TCC---T-GTCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCTGACAAACATGTATC-------------------TGTAT------------------CCATT--------CGAACGCGACAGA----TGTAG---------------------CA---------------------------------------AC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGCGCAAACACTTGACACGGGT--CCCTCCTGCTC-ATCTGAGT-CC----TGC-TCCTGTTGCC-CCCCG----------------------------C-------- |
| droTak1 |
scf7180000415383:93943-94195 + |
|
TGCTGT-----------T-G----TCC---T-GTCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATCCATCTATCC-------------AT------------------CCATT--------CGAACGCGAGAGA----TGTGG-------------------------------------------------------------CAG----------------------------------------------------------------------TGCCATCGCAATCCCTGAAATTGATTCCCTTTGTG-TTGCCAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGCGCAAACACTTGACACGGGT------GCTG-----------C-------TGC-TCCTTCTCC-----CAGTGAGTCCTCA-------------------------- |
| droEug1 |
scf7180000409787:1215169-1215400 + |
|
TTGTGC-----------T-G----TCC---T-GTCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCTGACAAACATGTATCC-------------------------------------------ATC--------CGAACGCGACAGA----TGTAG-------------------------------------------------------------CAC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGCGCAAACACTTGACATGGGT--C--TCCTCCCC-GTCT---------------------------------GAGTTCTCATTTT-------------G-------- |
| dm3 |
chr3R:24365461-24365694 - |
|
TTGTGC-----------T-G----TCC---T-GCCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATC-------------------CATAT------------------CCATT--------CAAACGCGACGGA----TGCGG-------------------------------------------------------------CAC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGT--C--TCTCCCTC-GCCT---------------------------------GAGTTCTCAT------------------------- |
| droSim2 |
3r:23743533-23743766 - |
dsi_29448 |
TTGTGC-----------T-G----TCC---T-GCCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATC-------------------CATAT------------------CCATC--------CAAACGCGACTGA----TGCGG-------------------------------------------------------------CAC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGT--C--TCCTCCTC-GCCT---------------------------------GAGTTCTCAT------------------------- |
| droSec2 |
scaffold_22:968624-968862 - |
|
TTGTGC-----------T-G----TCC---T-GCCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGTTGTGGTAAGCATCCGACAAACATGTATC-------------------CATAT------------------CCATC--------CAAACGCGACTGA----TGCGG-------------------------------------------------------------CAC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TTGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGT--C--TCCTCCTC-GCCT---------------------------------GAGTTCTCATCCT------------GC-------- |
| droYak3 |
3R:22131645-22131920 + |
|
TTGTGC-----------T-G----TCC---T-GCCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGCTGTGGTAAGCATCCGACAAACATGCAACCATGTATCC-CTGTATC--CATAT------------------CCATC--------CGAACGCGACGGA----TGCGG-------------------------------------------------------------CAC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TCGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGT--C--TCCTCCTCGGCCC---------------------------------GAGTTCTCATCCTCGTCCCCATCCTGCCACCCAGT |
| droEre2 |
scaffold_4820:3669950-3670196 + |
|
TTGTGC-----------T-G----TCC---T-GCCAGCA----AGGAGCC-AC-AAAATGGATTTATCGAGGCGGCTGTGGTAAGCATCCGACAAACATGTATCCATGTATC-----------CATAT------------------CCATC--------CAAACGCGACGGA----TGCGG-------------------------------------------------------------CAC----------------------------------------------------------------------------TGCAATCCCTGAAATTGATTCCCTTTGTG-TCGCTAGCAAATTACTGTGTCAATTGGAATTTTAATTCCGTTTGAACTACATGGGCAAACACTTGACACGGGT--C--TCCTCCTC-GCCA---------------------------------GAGTCCTCATCCT------------GC-------- |