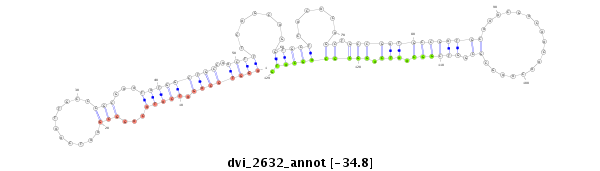
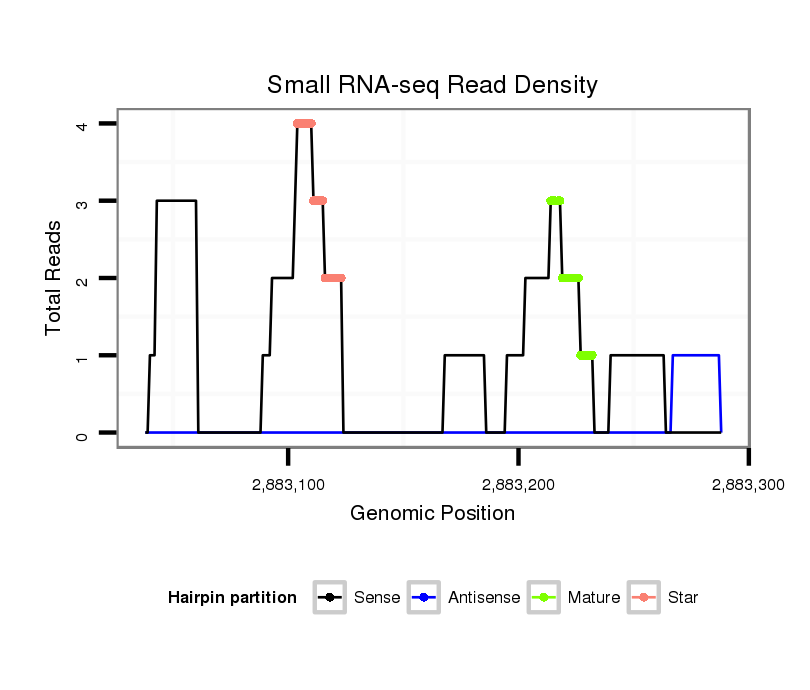
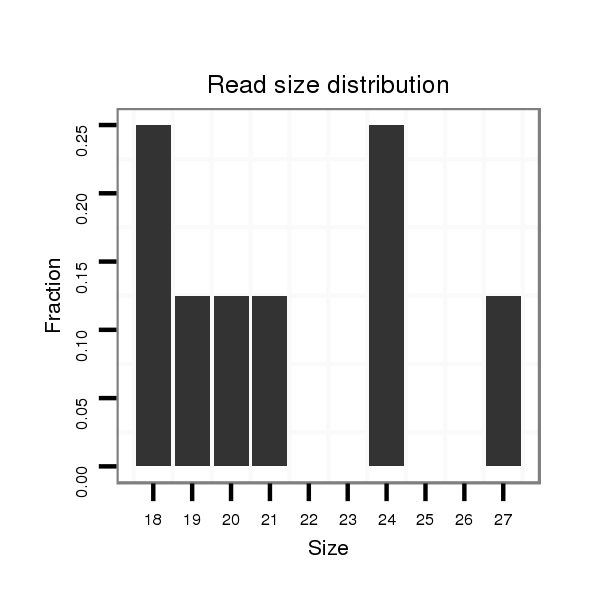
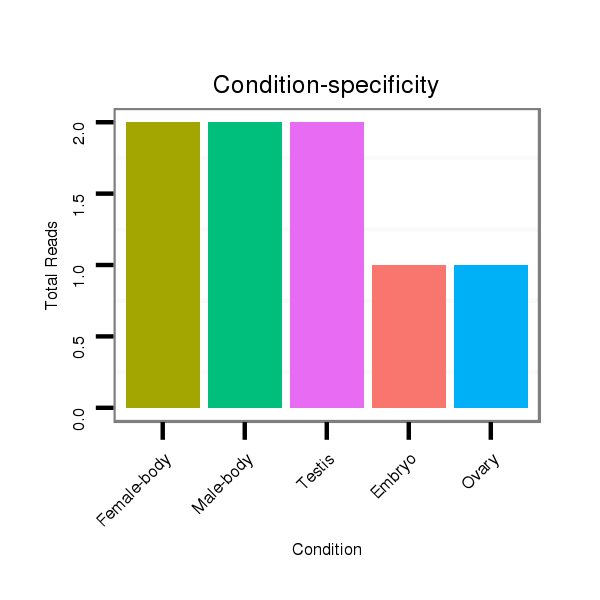
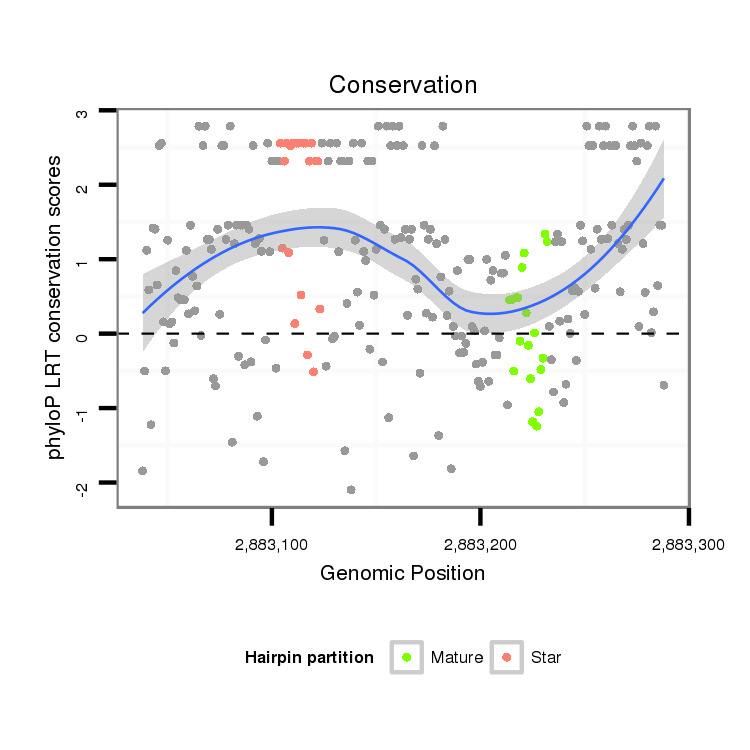
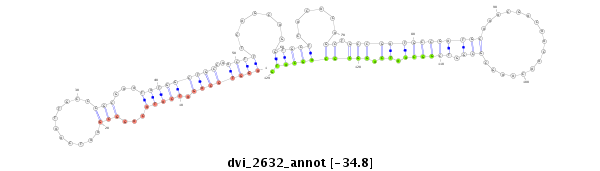
ID:dvi_2632 |
Coordinate:scaffold_12822:2883088-2883238 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.8 | -34.8 | -34.8 |
 |
 |
 |
exon [dvir_GLEANR_14467:2]; CDS [Dvir\GJ14523-cds]; intron [Dvir\GJ14523-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATCAAATGCAGCTGCAGTCGGGCAATCTGCAGGCGTTGCCCATACAAAATATACCCGGTCTGGGTCAGGTGCAGATCATACAGGCGAATCAATTGCCCGCGAATATGGCTGCCAACTTTCAGCAGGTGCTCACACAGCTGCCGATGCCGATGCAAACGACAACAACAAGCGCGGTCGGCAATCAGGCGCAGCATGCACCAGCACTGAGCATGCTGCCCAAGCAGGAGCCACAGAGTCCCACGCAGCTGATC ******************************************************************((((((((.(((((...(((...........)))...)))))))))..)))).......(((((......((.((((((((((((((.................)).)))))).))).)))))))))).******************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060663 160_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060665 9_females_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
V116 male body |
V053 head |
M047 female body |
SRR060666 160_males_carcasses_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....ATGCAGCTGCAGTCGGGC.................................................................................................................................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACTGAGCATGCTGCCCAAGCAGGA......................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GGCAATCAGGCGCAGCATG........................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGGTGCAGATCATACAGGCG..................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GACAACAACAAGCGCGGTCGGCAA...................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CACACAGCTGCCGATGCC....................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................CGGTCTGGGTCAGGTGCA.................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GGCGCGGTCGGCGATCAGGCGA.............................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAAATGCAGCTGCAGTCGGGC.................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CAGGTGCAGATCATACAGGCG..................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TACCCGGTCTGGGTCAGGTGCAGATCA............................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CAAGCGCGGTCGGCAATCAGGCGC.............................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CAAACGACAACAACAGGCGCG.............................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AAACGAGAACAACAAGCGC............................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ATGCTTCCCAAGCATGAGGC...................... | 20 | 3 | 20 | 0.25 | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................ACGCAAACGTCAACCACAAGCG................................................................................ | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................CAAGCTCGGCCGGCAGTCAG.................................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................TATGCTTCCCAAGCATGAG........................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GACCGGCAATCTGCTGGCG........................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TAGTTTACGTCGACGTCAGCCCGTTAGACGTCCGCAACGGGTATGTTTTATATGGGCCAGACCCAGTCCACGTCTAGTATGTCCGCTTAGTTAACGGGCGCTTATACCGACGGTTGAAAGTCGTCCACGAGTGTGTCGACGGCTACGGCTACGTTTGCTGTTGTTGTTCGCGCCAGCCGTTAGTCCGCGTCGTACGTGGTCGTGACTCGTACGACGGGTTCGTCCTCGGTGTCTCAGGGTGCGTCGACTAG
********************************************************((((((((.(((((...(((...........)))...)))))))))..)))).......(((((......((.((((((((((((((.................)).)))))).))).)))))))))).****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060682 9x140_0-2h_embryos_total |
V053 head |
SRR060686 Argx9_0-2h_embryos_total |
M028 head |
V047 embryo |
M027 male body |
M061 embryo |
SRR060687 9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR1106729 mixed whole adult body |
SRR060683 160_testes_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060673 9_ovaries_total |
SRR060669 160x9_females_carcasses_total |
SRR060681 Argx9_testes_total |
SRR060655 9x160_testes_total |
SRR060654 160x9_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060678 9x140_testes_total |
GSM1528803 follicle cells |
SRR060668 160x9_males_carcasses_total |
SRR060677 Argx9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................CGTCCACGAGAGGGTCGAC............................................................................................................... | 19 | 2 | 5 | 28.80 | 144 | 21 | 31 | 7 | 19 | 7 | 4 | 11 | 9 | 8 | 5 | 4 | 5 | 3 | 1 | 2 | 2 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TCGTCCACGAGAGGGTCGAC............................................................................................................... | 20 | 2 | 2 | 13.50 | 27 | 7 | 4 | 6 | 1 | 3 | 0 | 1 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GTCCACGAGAGGGTCGAC............................................................................................................... | 18 | 2 | 20 | 7.55 | 151 | 28 | 8 | 7 | 10 | 7 | 46 | 13 | 0 | 5 | 6 | 4 | 2 | 2 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 1 | 1 |
| .........................................................................................................................CGTCCACGAGAGGGTCGA................................................................................................................ | 18 | 2 | 9 | 4.00 | 36 | 12 | 0 | 6 | 3 | 1 | 0 | 3 | 1 | 2 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................TCGTCCACGAGAGGGTCGA................................................................................................................ | 19 | 2 | 3 | 3.33 | 10 | 3 | 1 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CGTCCACGAGAGGGTCGACT.............................................................................................................. | 20 | 3 | 20 | 1.25 | 25 | 2 | 9 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TTTGCTCTTGTTGTTCGCGCA............................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TCGTCCACGAGAGGGTCGACT.............................................................................................................. | 21 | 3 | 8 | 1.00 | 8 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GTCTCATGGTGCGTCGACGA. | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGTCTCAGGGTGCGTCGACTA. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GTCTCATGGTGCGTCGAC... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTCGTCCACGAGAGGGTCGAC............................................................................................................... | 21 | 3 | 8 | 0.25 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TGGTCCGCGTCATACGTGATC.................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................AGTATGACCGCTAAGTTAAG............................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTCGTCCACGAGAGGGTCGA................................................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CGTCCACGAGAGGGTCGAT............................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GTCCACGAGAGAGTCGAC............................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GTCCACGAGAGGGTCGA................................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12822:2883038-2883288 + | dvi_2632 | ATCA----------A-----------------------------A---------TGCAGCTGCA-GTCGGGCAATCTGCAGGCGTTGCCCATACAAAATATACC----CGGTCTGGGTCAGGTGCAGATCATACAGGCGAATCAATTGCCCGCGAATATGGCTGCCAACTTTCAGCAGGTGCTCACACAGCTGCCG---ATGCCG--------------------ATGCAAACGACAACAACAA---GCG----------------------------------CGGTCGGCAA------T--C------------AGG----------------------CGCAGCATGCAC---CA------GCACTGAGC----------ATGCTGCCCAAGCAGGAGCCACAGAGTCCCACGCAGCTGAT--------C |
| droMoj3 | scaffold_6540:7499437-7499714 - | GCCAAG---------CT------------------GCCGCTGCAA---------TGCAGCTGCA-GTCGGGTAATTTGCAGGCATTGCCCATACAGAATATACC----CGGCCTGGGGCAGGTGCAGATCATACAGGCGAATCAGTTGCCAGCAAATCTGGCGGCCAATTTCCAACAGGTGCTCACACAATTGCCC---ATGCCC--------------------ATGCAGACGACGACGGCAA-------------------------------------------TGGGTAA------T--CAGGCGCAGAAC---ATGC------------AGA----GCACAACTGCGG---CAACTTCCGCGCTGACC----------ATGCTGCCCAAGCAGGAGCCACAGAGTCCCACCCAATTGAT--------T | |
| droGri2 | scaffold_14906:2131439-2131737 + | GTCAAA---------ATGCGGCTGCTGCTGCTGCTGCTGCTGCAATGCAATCGCTGCAAACGA-------GCAATCTGCAAGCGCTGCCCATCCAAAACATACC----CGGTCTGGGTCAGGTGCAGATCATCCAAGCCAGTCAATTGCCCGCCAGCATGGCAGCCAATTTCCAGCAGGTGCTCACCCAACTCCCA---ATGCCC--------------------A------CGATGCAAGCCACAAGTAATGCTGCAGCGCCACAG---------------------------------------------AAC---TCGA------------A-------TAGCAATGCAGCTGCACCGGCTGCTATAACC----------ATGCTGCCCAAGCAGGAGCCACAGAGTCCCACGCAGCTAAT--------C | |
| droWil2 | scf2_1100000005674:975-1206 + | AGCA----------GTGGC---AGCG---------GG--TGCAGA---------GGC-----G-------GCAGTCTGCAGAGGTGGCCTGTAAGCGGCGGACACCGCAG--------------C---------------------------------------GGAGGGTACAGCAGGGGCGGGTGCGGCGGCAGCGGTTACAGCAGGGGCGAATATAGCCGCAGCGGGGCCGGCAGGGGCG---AGTA-----------------------------TGG------------CGGC------------------A-----------------------------GTTGGCA---CA------GCAGGAGCGAGTATGGCAGCAGTTGGCCCAGCAGGGGCAAGCATGTC---AGCAGTTGGCCCAGCATGG | |
| dp5 | 2:14583418-14583689 - | CACAGG------------------CC---------CCCACTGCCA---------TGCAGCTACCCG----CCGGGTTGCAGGCCCTGCCCATACAGAATATCCC----AGGATTGGGGCAGGTGCAGATCATCCAGGCGAATCAGTTGCCCGCCAACCTGCCGGCCAATTTCCAGCAGGTGTTGCATCAACTGCCACAAATGCAA-----------------------------------------------------------CAGGTTCATG---CCCAG------------GCGCCA------------GCACAGGCCCAGGGACAAGCTCAGA----CGACCAATGCAA---CA------TCTATAACG----------GTGATGCCCAAACAGGAGCCACAGAGTCCCACGCAGATGAT--------C | |
| droPer2 | scaffold_0:5836908-5837179 + | CACAGG------------------CC---------CCCACTGCCA---------TGCAGCTACCCG----CCGGGTTGCAGGCCCTGCCCATACAGAATATCCC----AGGATTGGGGCAGGTGCAGATCATCCAGGCGAATCAGTTGCCCGCCAATCTGCCGGCCAATTTCCAGCAGGTGTTGCATCAACTGCCACAAATGCAA-----------------------------------------------------------CAGGTTCATG---CCCAG------------GCGCCA------------GCACAGGCCCAGGGACAAGCTCAGA----CGACCAATGCAA---CA------TCTATAACG----------GTGATGCCCAAACAGGAGCCACAGAGTCCCACGCAGATGAT--------C | |
| droAna3 | scaffold_13340:22447374-22447600 + | T------------------------G---------CCCACGCCAC---------TTCAGCTGC-------CCAACCTGCAGGCCCTGCCCATCCAGAACATACC----CGGACTGGGCCAGGTGCAGATCATCCAAGCCAACCAGCTGCCACCGAATCTGCCCGCCAACTTGCAGCAGGTGCTTACCCAGCTGCCCTCAGTGCCT-----------------------------------------------------------CAGA---------TCCAGACTGGT------GGGC------------------AGTCGCA--------------TCTG-----------------------------CAA----------GTGCTGCCCAAGCAGGAGCCGCAGAGTCCCACGCAGATGAT--------C | |
| droBip1 | scf7180000396721:707600-707829 - | CTCCA------------------GTG---------CCCACCCCGC---------TACAGCTAC-------CCAACCTGCAGGCCCTGCCCATCCAAAACATTCC----CGGATTGGGCCAGGTGCAAATCATACAAGCCAACCAGCTGCCCCCGAATCTGCCCGCCAACTTACAGCAGGTGCTCACCCAACTGCCC---GTGCCG-----------------------------------------------------------CAGGTTCA---------GACTGGT------GGGC------------------AGTCGCA--------------CTTG-----------------------------CAG----------GTGATGCCGAAGCAGGAGCCGCAGAGTCCCACGCAGATGAT--------C | |
| droKik1 | scf7180000302778:471349-471620 - | CACCGACACCCGTACCCAC---CGTA---------CCCGCGCAGA---------TGCAGCTAC-------CCAATCTGCAGGCCCTGCCCATCCAGAATATCCC----CGGATTAGGTCAGGTGCAGATTATTCAGGCCAATCAACTGCCGCCTAATCTGCCGGCCAATTTCCAGCAGGTGCTCACTCAACTACCC---ATGGCC-----------------------------------------------------------CAGGCTCAGG---CGCAG------------GTGCAAGGCCATGTGCAGAAC---GCCCAGGGACAGAGTCA--GGGG-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACGCAGATGAT--------A | |
| droFic1 | scf7180000453912:1216765-1216999 - | TCCA----------ACTGC---AGTG---------CCTGCGCCCA---------TGCAGTTGC-------CCAATCTGCAGGCGTTGCCCATACAGAACATCCC----AAATTTGGGCCAAGTGCAAATTATCCACGCCAATCAGTTGCCGCCAAATTTGCCGGCCAACTTTCAGCAGGTGCTCACCCAACTGCCG---ATGTCG-----------------------------------------------------------CAGGCGCAGG---TACAG------------CAGC------------------CAACCCA--------------TGTG-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACGCAAATGAT--------A | |
| droEle1 | scf7180000491008:354680-354914 - | TCCA----------GCTGC---CGTG---------CCCACGCCGA---------TGCAACTGC-------CCAATCTCCAGGCGTTGCCCATTCAGAACATACC----GGGCTTGGGTCAAGTGCAAATCATCCACGCCAATCAGTTGCCCCCGAATCTGCCGGCCAACTTTCAGCAGGTCCTGACCCAACTGCCC---ATGTCA-----------------------------------------------------------CAGGCCCATG---TTCAG------------ACCC------------------AGGGCCA--------------AGTG-----------------------------CAA----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACGCATATGAT--------T | |
| droRho1 | scf7180000779477:128005-128239 - | TCCA----------ACTGC---AGTG---------CCCACGCCTA---------TGCAGTTGC-------CCAATCTGCAGGCGCTGCCCATCCAGAACATACC----TGGGCTGGGCCAAGTGCAAATCATCCATGCCAACCAGTTACCCCCAAATCTGCCGGGCAACTTTCAGCAGGTGCTGACCCAACTGCCC---ATGTCA-----------------------------------------------------------CAGGCCCATG---TTCAG------------AGCC------------------AGGGCCA--------------GGTG-----------------------------CAA----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACCCAGATAAT--------C | |
| droBia1 | scf7180000302136:402419-402652 - | CCCA-----------------------------------CGCCCA---------TGCAGTTGC-------CCAATCTTCAGGCGTTGCCCATCCAGAATATCCC----CGGGTTGGGTCAAGTGCAAATCATTCACGCCAACCAGTTGCCGCCAAATCTGCCGGCCAACTTCCAGCAGGTGCTTACCCAACTGCCG---ATGTCG-----------------------------------------------------------CAGGCTCAGG---TGCAA------------ACCC------ATGTGGCGAACCAGGCCCA--------------GGTG-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCGACGCAGATGAT--------T | |
| droTak1 | scf7180000415769:2400142-2400361 + | ACAA----------------------------------------A---------TGCAGCTGC-------CCAATCTGCAGGCTTTGCCCATTCAGAATATCCC----CGGTTTGGGTCAAGTGCAGATCATTCATGCCAACCAATTGCCGCCCAATCTGCCGGCCAATTTCCAGCAGGTACTCACCCAACTGCCC---ATGTCG-----------------------------------------------------------CAAGCTCAGGCGGTGCAA------------TCCC------------------AGGTGCA--------------AACC-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACGCAGATGAT--------C | |
| droEug1 | scf7180000409798:338240-338475 + | T------------------------G---------CCCACGCCAA---------TGCAGTTGC-------CTAACCTCCAGGCGCTTCCCATTCAAAACATCCC----CGGCTTGGGTCAAGTGCAAATCATTCACGCCAACCAGTTGCCGTCAAATCTACCGGCCAACTTCCAGCAGGTGCTTACACAACTGCCC---ATGTCT-----------------------------------------------------------CAGACTCAGG---TGCAA------------AACC------ATTTACCGACCCAGGGCCA--------------GGTG-----------------------------CAA----------GTTATGCCTAAGCAAGAGCCCCAGAGTCCGACGCAGATGAT--------C | |
| dm3 | chr3R:20050731-20050947 - | ACCA----------------------------------------A---------TGCAGATGC-------CCAATCTTCAGGCTTTACCCATCCAGAACATCCC----AGGCCTGGGTCAAGTGCAAATCATTCACGCCAATCAGCTGCCTCCTAATCTGCCAGCCAACTTCCAGCAAGTACTAACCCAACTACCC---ATGTCG-----------------------------------------------------------CATCCTCAAG---TGCAA------------ACTC------------------AGGGCCA--------------AGTT-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCAACGCAAATGAT--------C | |
| droSim2 | 3r:19582567-19582783 - | ACCA----------------------------------------A---------TGCAAATGC-------CCAATCTTCAGGCTCTGCCCATCCAGAACATCCC----AGGCCTGGGTCAAGTGCAAATCATTCACGCCAACCAGCTGCCTCCTAATCTGCCAGCCAACTTCCAGCAAGTACTAACCCAACTACCC---ATGTCG-----------------------------------------------------------CATCCTCAAG---TGCAA------------ACTC------------------AGGGCCA--------------AGTG-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCAACGCAAATGAT--------C | |
| droSec2 | scaffold_0:20383077-20383293 - | ACCA----------------------------------------A---------TGCAGATGC-------CCAATCTTCAGGCTCTACCCATCCAGAACATCCC----AGGCCTGGGTCAAGTGCAAATCATTCACGCCAATCAGATGCCTCCTAATCTGCCAGCCAACTTCCAGCAAGTACTAACCCAACTACCC---ATGTCG-----------------------------------------------------------CATCCTCAAG---TGCAA------------ACTC------------------AAGGCCA--------------AGTG-----------------------------CAA----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCAACGCAAATGAT--------C | |
| droYak3 | 3R:26456268-26456484 + | ACCA----------------------------------------A---------TGCAGATGC-------CTAATCTTCAGGCGCTGCCCATACAGAACATACC----GGGCTTGGGTCAAGTGCAAATCATTCACGCCAATCAGCTGCCGCCAAATCTGCCAGCCAACTTTCAGCAAGTACTTACCCAACTACCA---ATGCCG-----------------------------------------------------------CAGCCTCAGG---TGCAA------------ACTC------------------AGGGCCA--------------AGTG-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACGCAAATGAT--------C | |
| droEre2 | scaffold_4820:8011658-8011874 + | ACCA----------------------------------------A---------TGCAGATGT-------CCAATCTTCAGGCGCTGCCCATCCAGAATATCCC----AGGCTTGGGTCAAGTCCAAATCATTCACGCCAATCAGCTGCCTCCAAATCTGCCAGCCAACTTCCAGCAAGTACTTACCCAACTACCC---ATGTCG-----------------------------------------------------------CAGCCTCAGG---TGCAA------------ACTC------------------AGGGCCA--------------AGTG-----------------------------CAG----------GTGATGCCCAAGCAGGAGCCGCAGAGTCCCACGCAAATGAT--------C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:15 PM