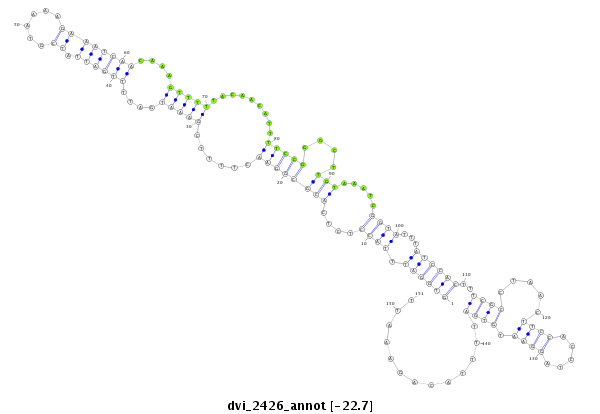
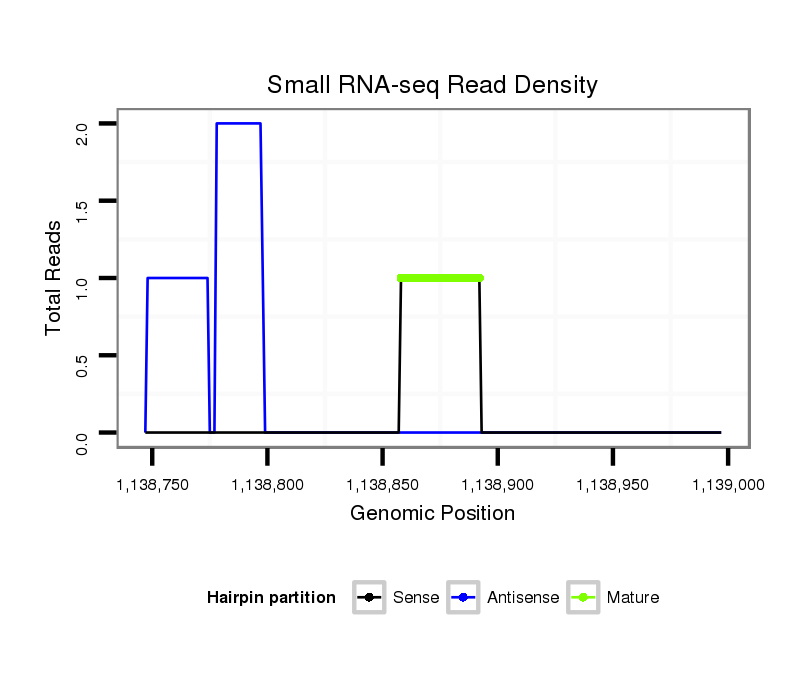
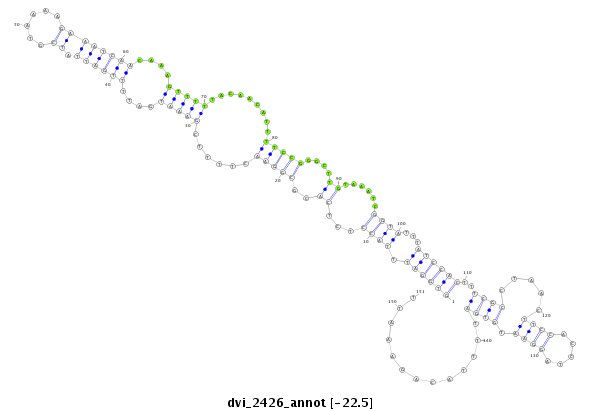
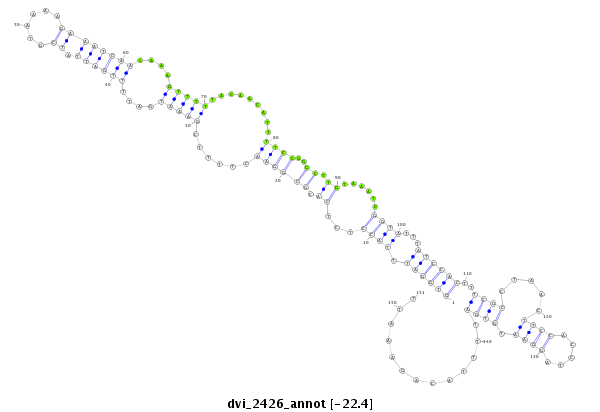
ID:dvi_2426 |
Coordinate:scaffold_12822:1138797-1138947 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -22.5 | -22.4 | -22.3 |
 |
 |
 |
exon [dvir_GLEANR_14381:1]; CDS [Dvir\GJ14436-cds]; intron [Dvir\GJ14436-in]
No Repeatable elements found
| -------------------###############################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACAAACTCAGTTTAAGCCTATGGCATTAATTCCAGTTTCCATCAAAATTCGTGGATTTACCTCTCACGCGGAACTTTTCGAAATGATTTTGATTATCGTAAAAGAAATCAACAAAGTTTTTACAACATTTTCCGGGCTTGTAAATCGGTATTTATCCACTTTCGCCTAACTTCCACCTAGGAATGTGATTTTACAGAAATTCACCTAACTTTCACCTAGGTTTGTGGAAAGAGTGAAAATCATTTTTCAAG **************************************************((((((.((((....((((((((......(((((....((((((.((......))))))))....))))).........)))))....))).....))))...))))))..((((.....((((.....)))).)))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M028 head |
SRR1106718 embryo_6-8h |
V047 embryo |
SRR060672 9x160_females_carcasses_total |
SRR060665 9_females_carcasses_total |
M027 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................CTAGGTTTGTGGAAAGAGAA................ | 20 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................CAAAGTTTTTACAACATTTTCCGGGCTTGTAAATC......................................................................................................... | 35 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................GTAAAAGAAAACAACAAGGTT..................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................TATGCGAGCTTGTAAATCGGT...................................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................ACGCGGACCTTTTCGAAC........................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................GCGGAACTGTTCGACAGGATT................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................CTACCAACTAGCAATGTGA............................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................ACTTCGTGGAATTACCTAT........................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
TGTTTGAGTCAAATTCGGATACCGTAATTAAGGTCAAAGGTAGTTTTAAGCACCTAAATGGAGAGTGCGCCTTGAAAAGCTTTACTAAAACTAATAGCATTTTCTTTAGTTGTTTCAAAAATGTTGTAAAAGGCCCGAACATTTAGCCATAAATAGGTGAAAGCGGATTGAAGGTGGATCCTTACACTAAAATGTCTTTAAGTGGATTGAAAGTGGATCCAAACACCTTTCTCACTTTTAGTAAAAAGTTC
**************************************************((((((.((((....((((((((......(((((....((((((.((......))))))))....))))).........)))))....))).....))))...))))))..((((.....((((.....)))).)))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060670 9_testes_total |
SRR060679 140x9_testes_total |
GSM1528803 follicle cells |
SRR060686 Argx9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
V116 male body |
SRR060661 160x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .GTTTGAGTCAAATTCGGATACCGTAAT............................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TGAAGGTATATCCTTACCCT............................................................... | 20 | 3 | 3 | 1.00 | 3 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ...............................GGTCAAAGGTAGTTTTAAGCA....................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................GGTCAAAGGTAGTTTTAAGC........................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................CATTTTCTTTAGTTG........................................................................................................................................... | 15 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................TTACTAACACTAAGAGCATG...................................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................GTAAAAGGCCCGATCAAGTA.......................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12822:1138747-1138997 + | dvi_2426 | ACAAACTCAGTTTAAGCCTATGGCATTAATTCCAGTTTCCATCAAAATTCGTGGATTTACCTCTCACGCGGAACTTTTCGAAATGATTTTGATTATCGTAAAAGAAATCAACAAAGTTTTTACAACATTTTCCGGGCTTGTAAATCGGTATTTATCCACTTTCGCCTAACTTCCACCTAGGAATGTGATTTTACAGAAATTCACCTAACTTTCACCTAGGTTTGTGGAAAGAGTGAAAATCATTTTTCAAG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 04:38 AM