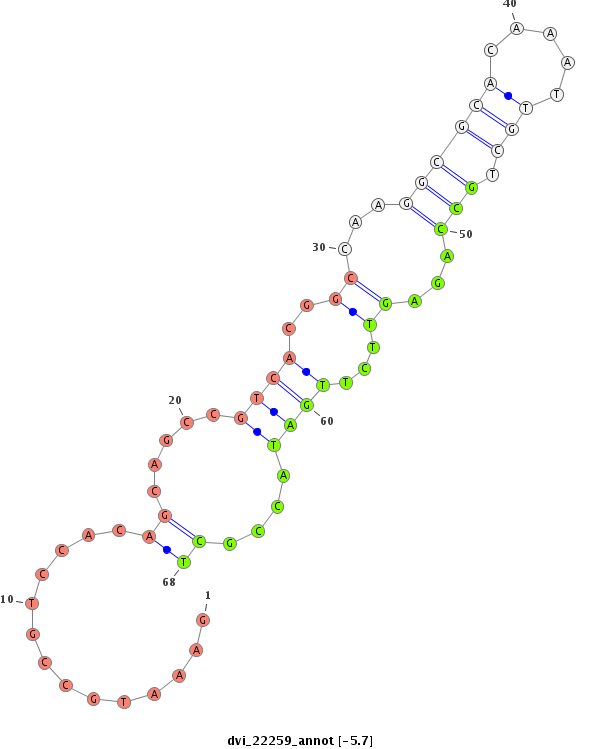
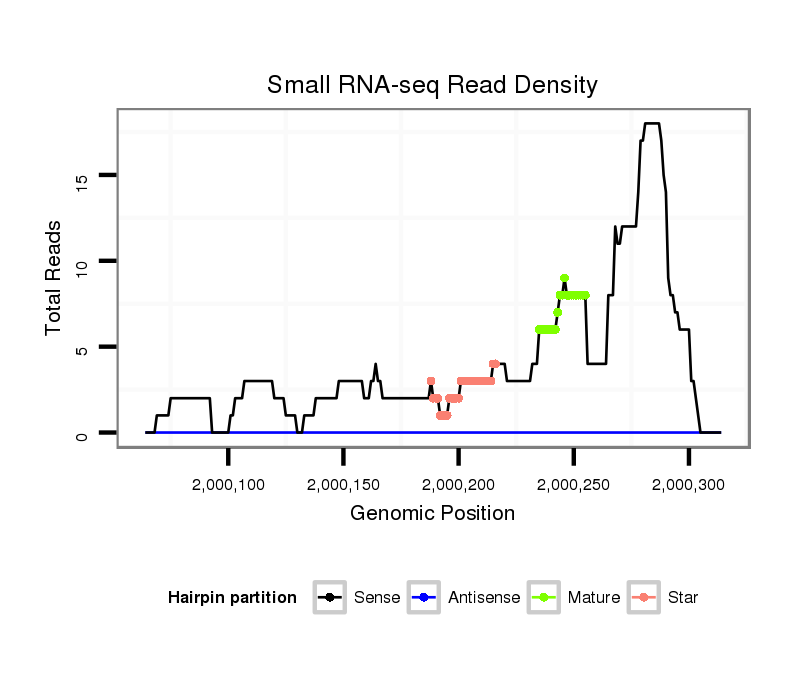
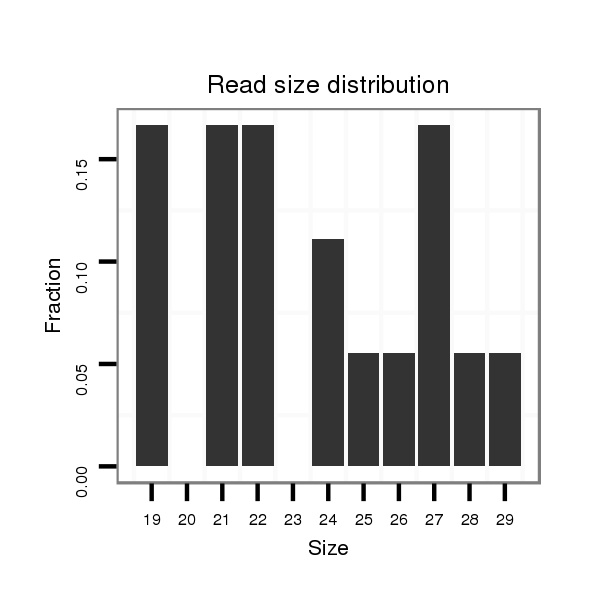
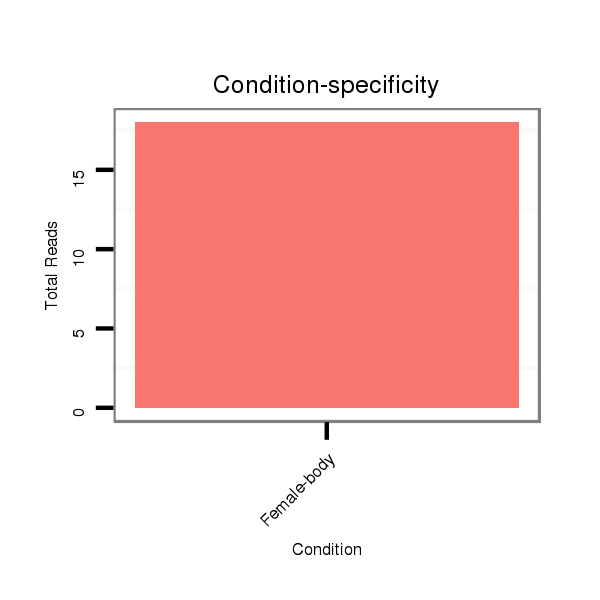
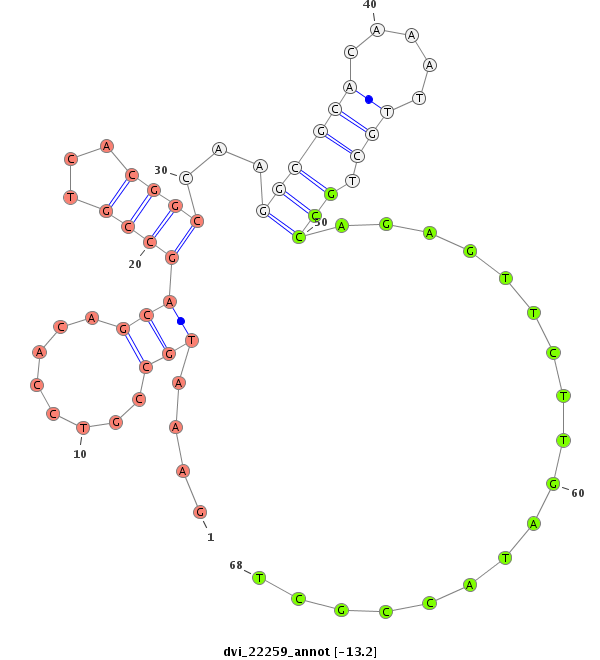
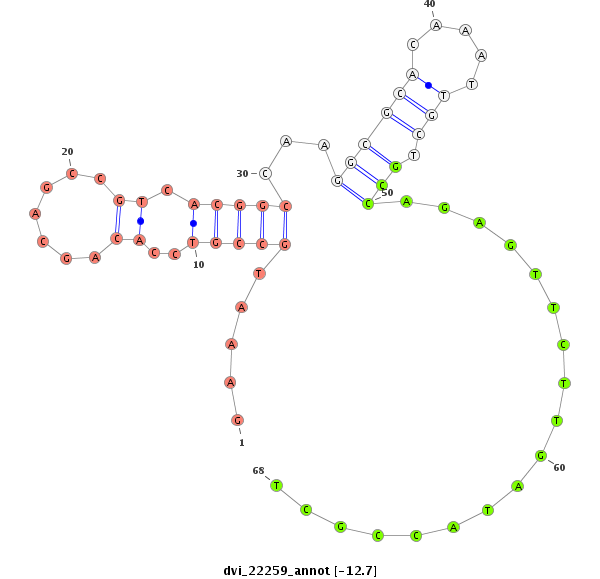
ID:dvi_22259 |
Coordinate:scaffold_13246:2000114-2000264 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.2 | -12.7 |
 |
 |
exon [dvir_GLEANR_16714:2]; CDS [Dvir\pdm2-cds]; intron [Dvir\pdm2-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGTGGCAACCGGCAGCCAGCCAGCGTTGGCACCCTTTGGCCTCGATCTCGGTCTGGGTCTGGGCCTGGGTTTGGGTCTCGGTCTGGGCCCGGAGCCGCAGACATGTCTGCCATATTTTGGCCTCGAAATGCCGTCCACAGCAGCCGTCACGGCCAAGGCGCACAAATTGCTGCCAGAGTTCTTGATACCGCTGCACGAAATCATGTTTCAACATCATTATCAGCAGCAGCAACAACAGCAGCAGCAGCAAC ****************************************************************************************************************************..............((.....((((..((...((((((.....))).)))...))...))))....))*********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V053 head |
SRR060667 160_females_carcasses_total |
M028 head |
SRR060665 9_females_carcasses_total |
V116 male body |
M027 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060683 160_testes_total |
SRR060684 140x9_0-2h_embryos_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................CATGTTTCAACATCATTATCAGCAGC........................ | 26 | 0 | 1 | 4.00 | 4 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GCCAGAGTTCTTGATACCGCT........................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CATGTTTCAACATCATTATCAGCA.......................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................ATTATCAGCAGCAGCAACAACA.............. | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GCTGCCAGAGTTCTTGATACCGCT........................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TCAACATCATTATCAGCAGCAGCAA................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................ATTATCAGCAGCAGCAACAACAGCA........... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GAAATGCCGTCCACAGCAGCCGTCACGGC.................................................................................................. | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CGAGATCATGTTGCAACCTCATT................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CAACCGGCAGCCAGCCAGCGTTGG.............................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................TCTTGATACCGCTGCACGAAAT.................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................CATTATCAGCAGCAGCAACAACAGCAG.......... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................GGGCCCGGAGCCGCAGACA.................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CAGCAGCCGTCACGGCCAAGGC............................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................CAGAGTTCTTGATACCGCT........................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GCAGCCAGCCAGCGTTGG.............................................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GTCCACAGCAGCCGTCACGGCCAAG.............................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GTTTCAACATCATTATCAGC........................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................GATCTCGGTCTGGGTCTG.............................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................CTTGATACCGCTGCACGAAATCATG.............................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GTTTCAACATCATTATCAGCAGC........................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GCACGAAATCATGTTCCAACATC.................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AGAGTTCTTGATACCGCTGCACGAAAT.................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................GCACAAATTGCTGCCAGAGTTCTT.................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................CCTCGATCTCGGTCTGGGTCTGGGCCT......................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AGACATGTCTGCCATATTTTGGCCTCG.............................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CATGTTTCAACATCATTATCAGCAGCA....................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GTTTCAACATCATTATCAGCAGCAGC..................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GCCAAGGCGCACAAATTGCTGC.............................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TTTGGGTCTCGGTCTGGGCCCGGAGC............................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GTCTCGGTCTGGGCCCGGAGCCGCAGA...................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................CGTCCACAGCTGCCGTCAC..................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GTTTCAACATCATTATCAGCAG......................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................CATTATCAGCAGCAGCAACAACA.............. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TGATACCGCTGCACGAAAT.................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................TATCAGCAGCAGCAACAACAGC............ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................GGCCTCGATCTCGGTCTGG................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................ACATGTCTGCCATATTTTGGCCTCGAAA........................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................CAAGGCGCACAAATTGCTGCC............................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................ACCCTGTGGCCTCGAACTTGG........................................................................................................................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CTCGGTTTCGACCCGGAGCC........................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GCAACCGGCAGCAAGCAAG.................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................AGCAACAACAGCAGCAGCAGCA.. | 22 | 0 | 20 | 0.15 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................CGAAATTCCGTCCCCTGCA............................................................................................................. | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TCAGCAGCAGCAACAACAGCAGC......... | 23 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AGCAGCAGCAACAACAGCAGCA........ | 22 | 0 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTCTGGGTCTGGTCCTGG....................................................................................................................................................................................... | 19 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................ATCAGCAGCAGCAACAACAGC............ | 21 | 0 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AGCAGCAGCAACAACAGCAGCAGC...... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GCAGCAGCAACAACAGCAGCA........ | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................AGCAGCAACAACAGCAGCA........ | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CAGCAGCAGCAACAACAGCAGCAGCA..... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................CAACAACAGCAGCAGCAGCA.. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................CAACAACAGCAGCAGCAGC... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................................AACAACAGCAGCAGCAGCA.. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................AGCAGCAACAACAGCAGCAGCAGCA.. | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CAGCAGCAGCAACAACAGCAGCA........ | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GCAGCAGCAACAACAGCAG.......... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................AGCAACAACAGCAGCAGCA..... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................AGCAGCAACAACAGCAGCAGCA..... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACACCGTTGGCCGTCGGTCGGTCGCAACCGTGGGAAACCGGAGCTAGAGCCAGACCCAGACCCGGACCCAAACCCAGAGCCAGACCCGGGCCTCGGCGTCTGTACAGACGGTATAAAACCGGAGCTTTACGGCAGGTGTCGTCGGCAGTGCCGGTTCCGCGTGTTTAACGACGGTCTCAAGAACTATGGCGACGTGCTTTAGTACAAAGTTGTAGTAATAGTCGTCGTCGTTGTTGTCGTCGTCGTCGTTG
***********************************************************..............((.....((((..((...((((((.....))).)))...))...))))....))**************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060654 160x9_ovaries_total |
SRR060658 140_ovaries_total |
SRR060666 160_males_carcasses_total |
V116 male body |
M027 male body |
SRR060675 140x9_ovaries_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................AGCACTATGGCGAAGTGCT..................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TAGTAGAAAGTAGTAGTAATAG.............................. | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................AAGTCGTAGGAATAGTCGTC......................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GGTGTCGTAGGCATTGCTGG................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCGTCGTTGTTGTCGTC.......... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................AATAGTCGTCGTTGTTGTTGT.............. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................TGTCGGCGACAGTGCGGGTT............................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13246:2000064-2000314 + | dvi_22259 | TGT-G--GCA---ACCGGCA-G------------------CCAGCCAG--CGT-------TGG--CA----CCCTTTGGCCTCGATCTCGGTCTGGGTCTGGGCCTGGGT-TTGGGTCTCGGTC--------------------------------------------------------------------------------T----GGG---------------------CCCGGAGCCG--------------------------------------------CAGACATGTCTGCC---A-----------------TATTTTGGCCTCGAAATGCCGTC----------------------------------------------------------------------------------------------CACAG----------------CAGCCGTCACGGCCAAGGCGCA--CAAATTGCTGCCAGAGTTCTTGATACCGCTGCACGAAATCAT-GTTTC---A------ACATCATTATCAGC---AGCAGCAACAACA-GCAGCAGCAGCAAC |
| droMoj3 | scaffold_6500:3966902-3967131 - | TGT-G--GCA---GCTAGCA-G------------------TCAGGCGG--CGT-------TGC--CA----CCCTTTGGCGTCGGGCTCGGACTGGGTGTGGGCGTGGGC----------------------------------------------------------------------------------------------------------------------GCGGGCCTGGAGCCG--------------------------------------------CAGACATGTCTGCC---A-----------------TATTTTGGCTACGAAA--------------------CAG---------------------------------------------------------------------------------------------------CAGCCGTCACGGCGAAAGCGCA--CAAATTGCTGCCAGAGTTTTTGATACCATTGCATGAAATTAT-GTTTC---A------ACATCATTATCAAC---AACAACAACAACA-ACAACAACAGCAGC | |
| droGri2 | scaffold_15252:15279405-15279719 - | CGT-G--CCA---CCAACCACAACAACAACGACAACAGCAGCAA---------------------CA----ACATTTGGCGTCGATGTTGGCATGGGTTTGAATTTGGGC-TTGGGCTTAGGAGTGGGCGTGGCTGGCGTTGGGGTT------------------------------------------------GGGGGTGGC------------------------GGTGGA-----GT------GGGAGTTGCGGGGGAGTCGCC------------------GCAGACATGTCTGCC---A-----------------TATTTTGGCATGGAAATGG--AAATG---C-----------------------------------------------------------CGT---------------C---------AACAA----------------CAACCGTCTCCGCCAAAGCACA--CAAATTGCTGCCCGAATTTCTGATACCGTTGCATGAAATCAT-GTTTC---A------ACATCATTATCAAC---AGCAACGACAACAAGTTGTGGCAGCAAC | |
| droWil2 | scf2_1100000004521:10531964-10532187 + | TAT-A--ACC-----GTGC--A------------------ACAGCT--GATGT----------TG--------------------------------------------------------------------------------------------------------C------------------CGTTTC----------TAAGAGA-------------------TGGC--------------------GGAGCCACAGCAGATGGAGCCGGGGGTATAGGGCAGACATGTCTGCC---A-----------------TATTTCGGTTCGACGA---CAACATC---TGTCAATTC-----------------------------------------------------------------------------------------------------------------AAAGCTGGA--GAAATCCCTGCCAGACTTTCTAATGCCATTGCATGAAATTAT-GTTCC---A------CCATCATCATCATCATCAGCATCATCAGCA-TGCCCAAGAATATC | |
| dp5 | 4_group3:3421379-3421626 + | GGC-AGC--A---ACAGGGACA------------------TCAACAAGGATAT----------GGC-----------------------------------------------------------------------------------------------------------ACAC---ATGACTTCCGC-TC----------TG---GAGGGAGGAGCCATG-GGGGATGGC-----------------------------------------------------CAGACATGTCTGCTACCGGCCCCA--------TCATATTTTGGCATGGAAACACTGGCATC---TGTCAAGCAGCAG--------------------------------------CAGCAGCAGC------------ATC-----------------AG------------CA-------------------TCA--GAAATCCCTGCCAGACTTTCTAATACCATTGCATGAAATTAT-GTTCC---A------ACAACATCATTATC---AGCAG--AGAACA-GCAGCAGCAGCGGC | |
| droPer2 | scaffold_1:4897978-4898224 + | GC--AGC--A---ACAGGGACA------------------TCAACAAGGATAT----------GGC-----------------------------------------------------------------------------------------------------------ACAC---ATGACTTCCGC-TC----------TG---GAGGGAGGAGCCATG-GGGGATGGC-----------------------------------------------------CAGACATGTCTGCTGCCGGCCCCA--------TCATATTTTGGCATGGAAACACTGTCATC---TGTCAAGCAGCAG--------------------------------------CAGCAGCAGC------------ATC-----------------AG------------CA-------------------TCA--GAAATCCCTGCCAGACTTTCTAATACCATTGCATGAAATTAT-GTTCC---A------ACAACATCATTATC---AGCAG--AGAACA-GCAGCAGCAGCGGC | |
| droAna3 | scaffold_13340:182155-182368 + | GTC-C--GCA---GCCTCT----------------------------------------------------------------------------GTCTCAGCCTCAGTC-TCTGTCTCAGTCG------------CAGCCAAAGTCGAAGTCCTAGTTTCTGCCGCCG---------------------T-CG------------------------------------CAGT-------------CAGCAGCGCATCCACAACACCCAAAAC-------------CAACCACCGCCGCC---A-----------------CACACCTCCACCGAAG----------------------------------------------------------------------------------------------------------------------------------------------CA--G--ACGTCAGCGAACCCTGCCAACACCACCCCCC-----------------A------GCAACATCG-CAGC---AGCATCAAGAACA-ACAGCAGCAGCAAC | |
| droBip1 | scf7180000396580:796750-797029 + | TGG---C--A---GCGGGGAGC------------------TGGCGAAAGATGT----------TG--------------------------------------------------------------------------------------------------------CCCCGCAG---ATGACTTCCGCATGTCTC------TACCCGA-----------TG-GAGGATGCC-----------------------------------------------------CAGACATGTCTGCC---GGCC-----------TCGTATTTTGGCATGGAAACGCTGGCATC---CATCAAACAC--------------------CAACAACTCCAGCAACAACTACAGCAACGCCATCAGAACCAGCACCAGCAACAGCAGC-----AGCACCAGCCAACGAG-------------------ACA--GAAATCCCTGCCAGACTTTCTAATACCTCTGCACGAAATTAT-GTTCC---AACAGCAACAACACCATTACC---AACAA----------------------- | |
| droKik1 | scf7180000302472:1067100-1067324 + | CGT-G--G-----ACAGCGGCA------------------TCAACAAGGATGT-------CGT--CG-C---CGT-----------------------------------------------------------------------------------------------TCCGCAGATGATGACTTCCGCCTGTAGCCG------CCTGC------TGCCGCCGGAGGATGGT---CAGCAG--------------------------------------------CAGACATGTCTGCC---GGTC-----------TCATATTTTGGCATGGAAACGCTGGCTTCAACGGTCAAGCAG-------------------------------------------------------------------------------------------------CG-------------------CCA--GAAATCCCTGCCAGACTTTCTAATACCGCTGCACGAAATTAT-GTTCC---A------ACAACATCATTACC---AGCA------------------------ | |
| droFic1 | scf7180000452061:10355-10542 + | GG---------------GGAAA------------------TCAACAAGGATGT----------CG---C---CA-----------------------------------------------------------------------------------------------TCCAGCAG---ATGACTTCCTCATGTGG------C------------------------GATGGC-----------------------------------------------------CAGACATGTCTGCC---ACTC--------GCCTCGTATTTTGGCATGGATACGCTGGCATC---TGTCAAACAG-------------------------------------------------------------------------------------------------CG-------------------GCA--GAAATCCCTGCCAGACTTTCTAATACCGCTGCACGAAATTAT-GTTCC---A------ACAACATCATTACC---AGCA------------------------ | |
| droEle1 | scf7180000491282:144293-144483 - | GCA---CT---------------------------------CA----C--TGT----------CG---CCGCCCCACAGTGTGGGTGTGGGTGTGGGTGTGGGTGT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGCGTTGGCGTGGGCGTGGGCGTGGC---TGGCGAGGAT-----------------------------------------------------------------C-----------------AG------------CA-------------------CCGCAGAGCCTCGCTGCAGGGTGTCCAGGAGAAGCTCACCCAGCTGCAGATGCAACA------GCAACATCACCAGC---AGCAGCAGCAGCA-TCAACAGCAGCAAC | |
| droRho1 | scf7180000779506:63634-63850 + | GCC-G--C-C-G-TCGGCG----------------------------------------------------GACTCGGGCATTGGGATGGACCTGGGCCTGGGCGTGGGC-ATGGG------------------------------------CATGGGCATGGTCATGG---------------------------------------------------------------------------------------------------------------------------GCGTGGGCA---A-----------------CAAGCATGCTTCGAAG--------------------CAGCAGCCGCCTTTACCGGTGGTCAACTGCAACAACAATAACAACGGCAGCGGC--------------------------------------------------------------------------AATAGCGTTAGGAGCTGCAA-------------------------C---A------ACGGCAGCAGCAGC---AGCAGCAGCAACA-GCCACAGCAGCAGC | |
| droBia1 | scf7180000301760:2808298-2808516 + | CGG---C--G---GCCGCGGCG------------------GCAGCCGTTGCCTCGTCCGTAGC--CA-----CCTCCGTCTCCGTGTCCGTTTCCGGCTCGGGAGCGGGCTCCGCATCGGGATC--------------------------------------------------------------------------------G----GGT---------------------TCCGGAGCCGCAGCCAGTGC----------------------------------CTCAGCCAGCGCAG---CGAC-----------T--------------GGAG--------------------CAG---------------------------------------------------------------------------------------------------CAGCG---------------------------GTCCTGAGCCTGGAGAACAGCTTCACCCAGCCAG-GC-----------------AACCCCCACC---AGCAACAGCAGCC-GCAGCAGCAGCAAC | |
| droTak1 | scf7180000413103:22562-22818 + | TGT-T--GCC---GCCGG--CG------------------ATAATATAAGCGT-------GAG--CAAT--CCCTTCG---AGGATCCTGGTCCTGGCCCAGGTCCAGGT-GCTGGTCCCGGTG------------GTGCTGGAGCTGGAGCTGTT---------------------------GC--------------------------------TGCTGTG-GGCGGTGGTCCG--------------------------------------------------CAACCACATCCACC---GCCCCCACATTCACCG--CA---CACCGCAGCCCAACAGGCGGC---CGGCCAACAGCAG--------------------------------------CAGCAGCAGC------------ATC---------------------------------------------------------------------------CTCAGCATCAGCATCCT------------------------GGGCTGCCGGGGC---C---ACCGCCGCC-ACAGCAGCAACAGC | |
| droEug1 | scf7180000409122:1063843-1064034 + | AATCAGC--A---GCGGGGAAA------------------TCAACAAGGATGT----------TG--------------------------------------------------------------------------------------------------------GCCAACAG---ATGACTTCCGCATGTGG------C------------------------GATGGC-----------------------------------------------------CAGACATGTCTGCC---AGTC-----------TCGTATTTTGGCATGGAGACGCTGGCATC---TGTCAAACAG-------------------------------------------------------------------------------------------------AG-------------------ACA--GAAATCCCTGCCAGACTTTCTAATACCGTTGCACGAAATTAT-GTTCC---A------ACAACATCATTACC---AGCA------------------------ | |
| dm3 | chr2L:12672902-12673085 + | C-----------------------------------------AACAAGGATGT----------TA--------------------------------------------------------------------------------------------------------GCCAGCAG---ATGACTTCCGCATGTGGCGGTAGC------------------------GATGGC-----------------------------------------------------CAGACATGTCTGCC---AGTC-----------TCGTATTTTGGCATGGAGACGCTGGCATC---TGTCAAACAG-------------------------------------------------------------------------------------------------AG-------------------ACA--GAAATCCCTGCCAGACTTTCTAATACCACTGCACGAAATTAT-GTTCC---A---GCAACAACATCATTACC---AGCAA----------------------- | |
| droSim2 | 2l:12321902-12322100 + | CGG---C--A---GCGGGGAAA------------------TCAACAAGGATGT----------TG--------------------------------------------------------------------------------------------------------GCCAGCAG---ATGACTTCCGCATGTGGCGGTGGC------------------------GATGGC-----------------------------------------------------CAGACATGTCTGCC---AGTC-----------TCGTATTTTGGCATGGAGACGCTGGCATC---TGTCAAACAG-------------------------------------------------------------------------------------------------AG-------------------ACA--GAAATCCCTGCCAGACTTTCTAATACCGCTGCACGAAATTAT-GTTCC---A---GCAACAACATCATTACC---AGCAA----------------------- | |
| droSec2 | scaffold_16:857125-857324 + | CAG---C--GGGTGCGGGGAAA------------------TCAACAAGGATGT----------TG--------------------------------------------------------------------------------------------------------GCCAGCAG---ATGACTTCCGCATGTGGCGGTGGC------------------------GATGGC-----------------------------------------------------CAGACATGTCTGCC---AGTC-----------TCGTATTTTGGCATGGAGACGCTGGCATC---TGTCAAACAG-------------------------------------------------------------------------------------------------AG-------------------ACA--GAAATCCCTGCCAGACTTTCTAATACCGCTGCACGAAATTAT-GTTCC---A---GCAACAACATCATTACC---AGC------------------------- | |
| droYak3 | 2L:4626668-4626884 - | CAA---C--C--------------------------------AGACAG--AGC-------ACG--AG----CCTTTTGGCATGAGCCTC-CTCCAAGTCTGAATCTGAAT-CTGAATCTCAG--------------------------------------------------------------------TCTGGGGCTGATGC------------------------TGAGGC-----------------------------------------------------TGAGGCTGAGTCAG---GGGC-----------TCGAGCTCTGACATGGGTT----GTTCTT-------------------------------------------------------------------------------------------------------------------------------------T--GGACTCCCTGCCA--CCTTGGAAT--AGGCAAAATTAACTAA-ATTAC---A------ACATCAACATCAAC---GGCAGCAACTGCA-GCAACAGCAGCGAA | |
| droEre2 | scaffold_4929:12749888-12750071 - | GG-----------------AAA------------------TCAACAAGGATGT----------TG--------------------------------------------------------------------------------------------------------GCCAGCAG---ATGACTTCCGCATGTGG------C------------------------GATGGC-----------------------------------------------------CAGACATGTCTGCC---AGTC-----------TCGTATTTTGGCATGGAGACGCTGGCATC---TATCAAACAG-------------------------------------------------------------------------------------------------AG-------------------ACA--GAAATCCCTGCCAGACTTTCTAATACCGCTGCACGAAATTAT-GTTCC---A---GCAACAACATCATTACC---AGCAA----------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:55 AM