
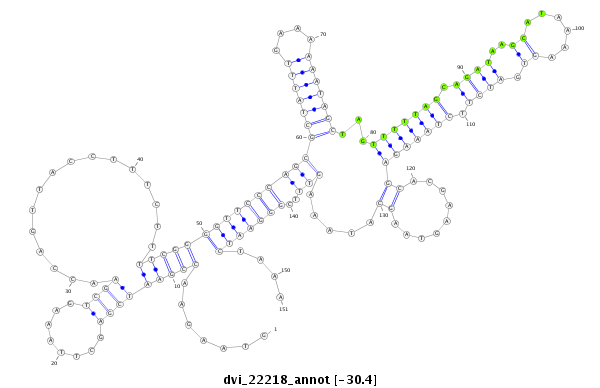
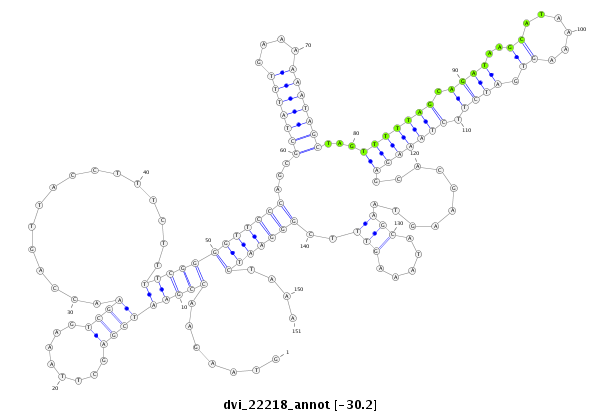
ID:dvi_22218 |
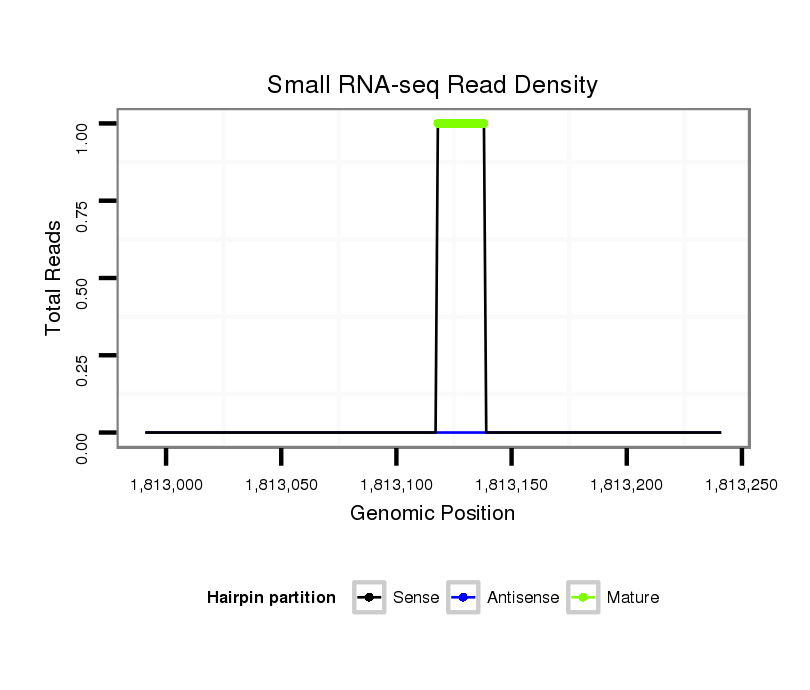
Coordinate:scaffold_13246:1813041-1813191 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -30.4 | -30.4 | -30.2 |
 |
 |
 |
CDS [Dvir\GJ16270-cds]; exon [dvir_GLEANR_16710:1]; intron [Dvir\GJ16270-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCCAAATTTGGTCAGAGCTCGAATGAAAAACCAGATATGGAAAAGCAGCTGTAAGAACCGAATCGAGCTTAAAGTCGAACCAGTTACCTTTCTTTTCGGGGTTCCCAGCGCTATTTGAAAAAATAGCTAGTTTTTAGCAGATAAGCATAAAAGTGATCTTCTAAAGAGCACGAAGTAAGCATAAAGTTTCGGGAATCTAAAAAGAAATTTCGTTTGCATAGATTTTTTACTTTATCTTTATATTGATCAAA **************************************************.......(((((((((........))))................)))))(((((((...(((((((....)))))))...(((((((.((((..((......)).)))).))))))).........((((.....)))).)))))))....************************************************** |
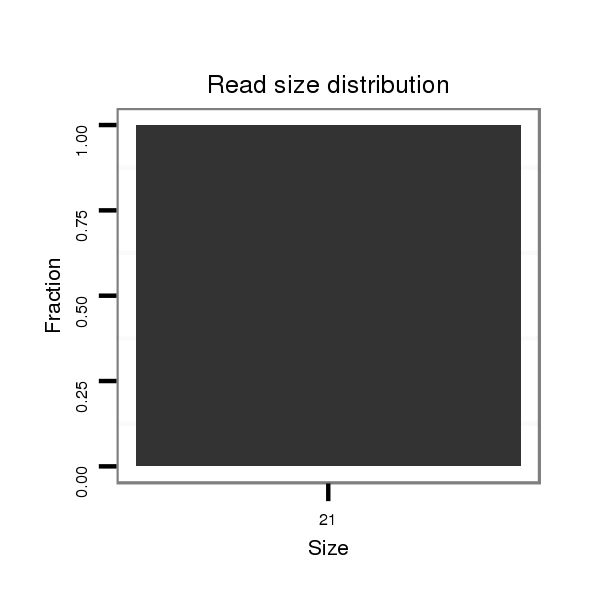
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
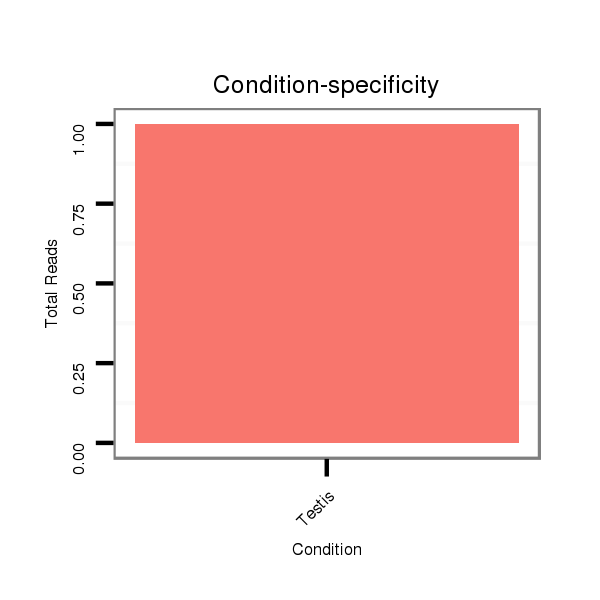
SRR060678 9x140_testes_total |
V116 male body |
SRR060684 140x9_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060687 9_0-2h_embryos_total |
M047 female body |
SRR060689 160x9_testes_total |
M028 head |
SRR060658 140_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
V053 head |
SRR060660 Argentina_ovaries_total |
SRR060669 160x9_females_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060679 140x9_testes_total |
M027 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................TTTACTGTAGCTTGATATTGA..... | 21 | 3 | 13 | 9.00 | 117 | 50 | 22 | 2 | 11 | 11 | 0 | 5 | 4 | 2 | 2 | 0 | 2 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TAGTTTTTAGCAGATAAGCAT....................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................CCCAGCGCCATCTGAAAAAA................................................................................................................................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TTACTGTATCTTGATATTGA..... | 20 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CCAGCGCCATCTGAAAAA................................................................................................................................. | 18 | 2 | 13 | 0.23 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................AAGTCGAACGAGTTTCCGTT................................................................................................................................................................ | 20 | 3 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ........................GAAGAACCAGATTTGGAA................................................................................................................................................................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GTTATAGAAAAGCAGCTCTAA..................................................................................................................................................................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TTTGGGAATGCAGCTGTAAG.................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CCAGCGCCATCTGAAAAAAA............................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................AATGAGATATGGAAAAGC............................................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................AAGTCGAACGAGTTTCCGT................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGGTTTAAACCAGTCTCGAGCTTACTTTTTGGTCTATACCTTTTCGTCGACATTCTTGGCTTAGCTCGAATTTCAGCTTGGTCAATGGAAAGAAAAGCCCCAAGGGTCGCGATAAACTTTTTTATCGATCAAAAATCGTCTATTCGTATTTTCACTAGAAGATTTCTCGTGCTTCATTCGTATTTCAAAGCCCTTAGATTTTTCTTTAAAGCAAACGTATCTAAAAAATGAAATAGAAATATAACTAGTTT
**************************************************.......(((((((((........))))................)))))(((((((...(((((((....)))))))...(((((((.((((..((......)).)))).))))))).........((((.....)))).)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
M027 male body |
SRR060666 160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
M028 head |
SRR060665 9_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................TAGCTCAAAGCCCTTAGAATTT................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TATTTCCAAGCCCTTAGAATTT................................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TAGTTCCAAGGCCTTAGATTT.................................................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TATTTCCAAGGCCTTAGAATTT................................................. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ATTTCCAAGCCCTTAGAATTG................................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ATTTCCATGCCCTTAGAATTT................................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TATTTCCAAGTCCTTAGAATTT................................................. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTCCAAGCCCTTAGAATTT................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TAGTTCCAAGCCCTTAGAATTT................................................. | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGGAAAGCCCCAAGGCTG............................................................................................................................................... | 18 | 3 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TCCAAGGCCTTAGAATTT................................................. | 18 | 3 | 20 | 0.25 | 5 | 0 | 0 | 0 | 3 | 1 | 0 | 1 |
| .....................................................................................TGCAAGGTAAAGCCCCAAGG.................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTCCAAGCCCTTAGAATTG................................................. | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTCCAAGGCCTTAGAATTT................................................. | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................AAGAGGAAAGCCCCAAGG.................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
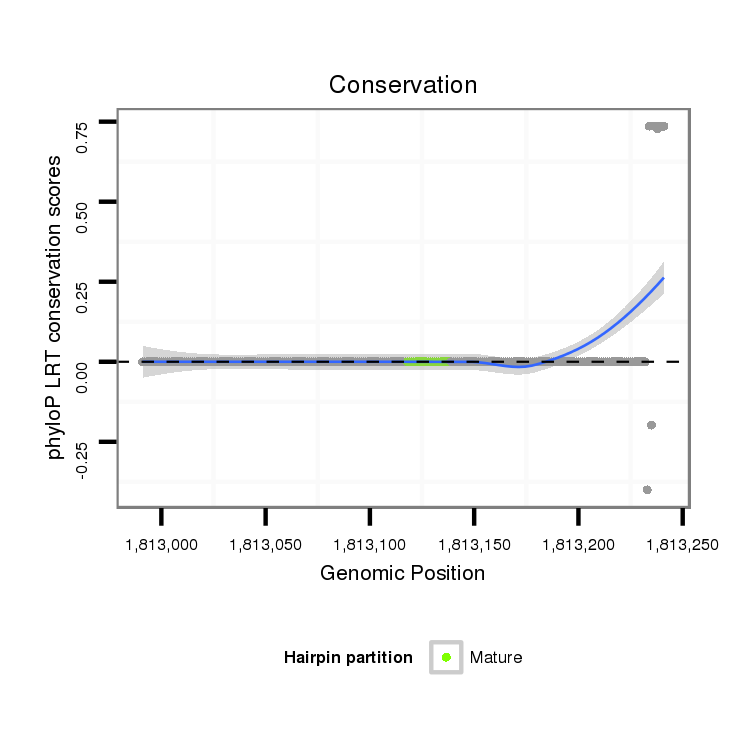
| droVir3 | scaffold_13246:1812991-1813241 + | dvi_22218 | GCCAAATTTGGTCAGAGCTCGAATGAAAAACCAGATATGGAAAAGCAGCTGTAAGAACCGAATCGAGCTTAAAGTCGAACCAGTTACCTTTCTTTTCGGGGTTCCCAGCGCTATTTGAAAAAATAGCTAGTTTTTAGCAGATAAGCATAAAAGTGATCTTCTAAAGAGCACGAAGTAAGCATAAAGTTTCGGGAATCTAAAAAGAAATTTCGTTTGCATAGATTTTTTACTTTATCTTTATATTGATCAAA |
| droGri2 | scaffold_15252:15449726-15449726 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-------- | |
| droAna3 | scaffold_12916:12852619-12852627 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAATCAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
Generated: 05/17/2015 at 04:53 AM