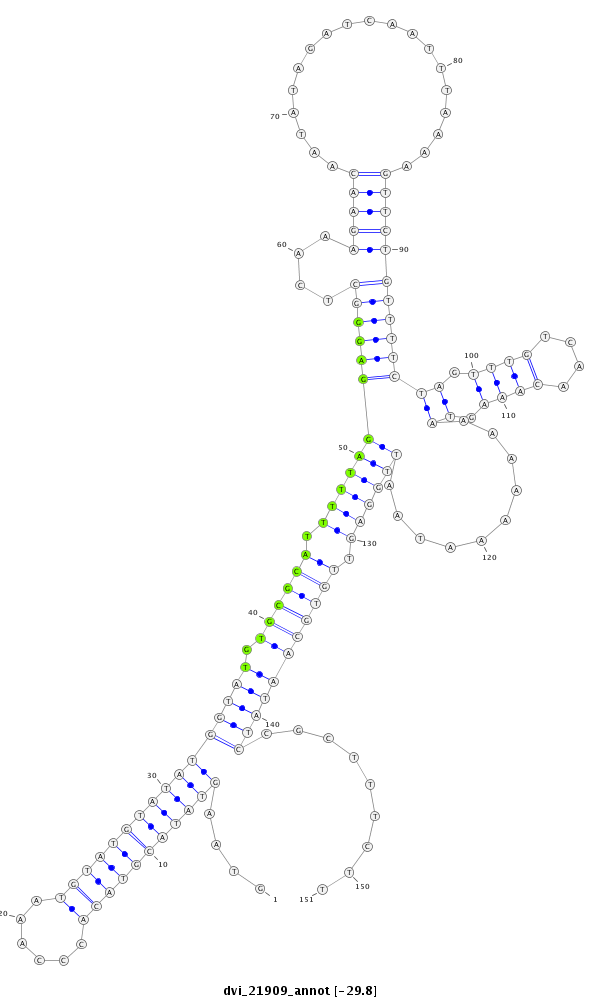
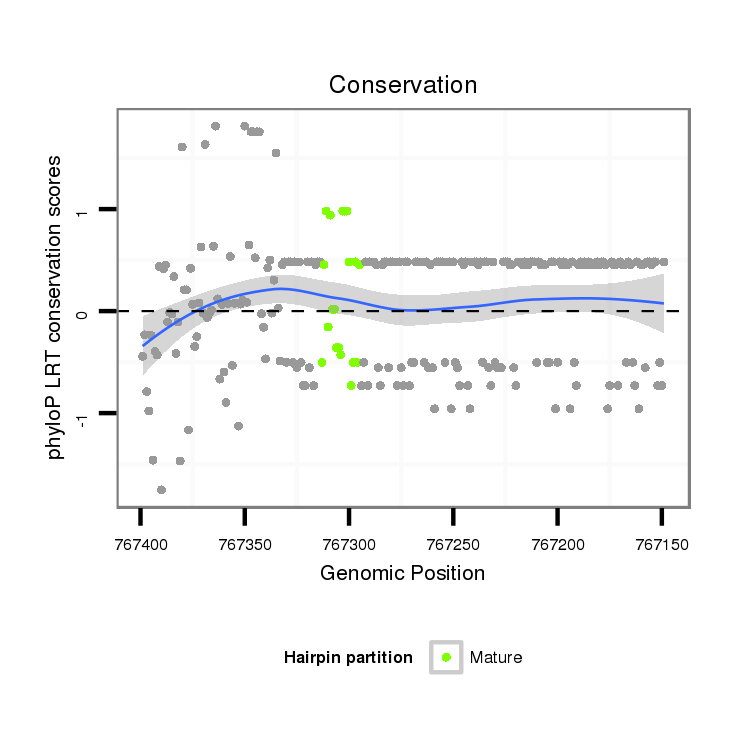
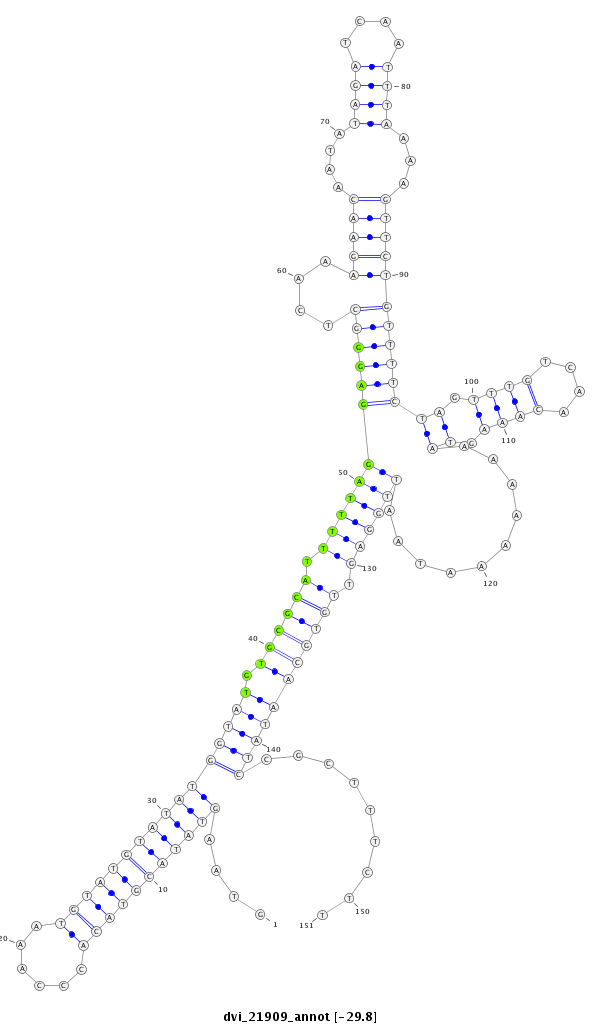
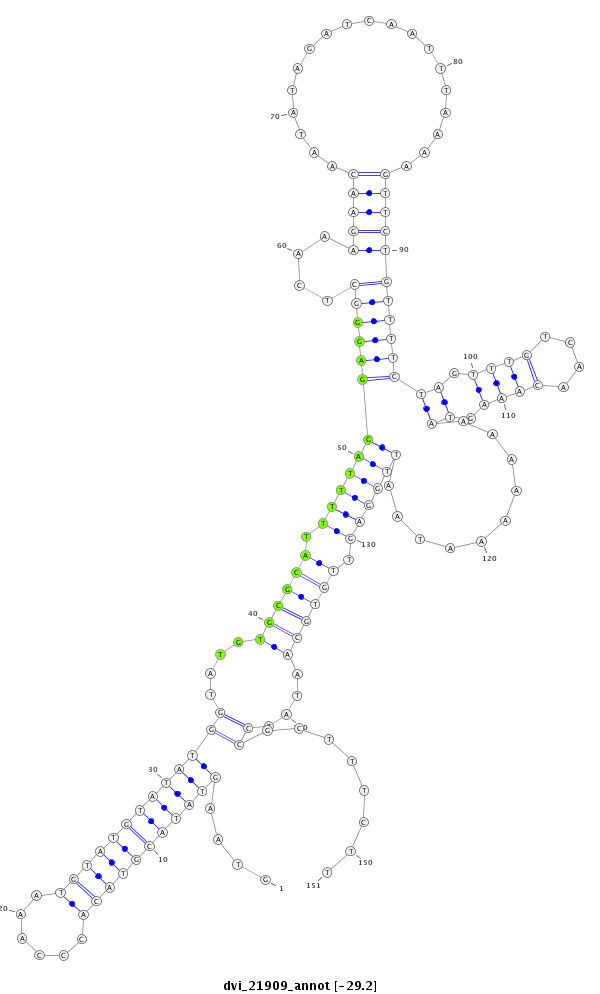
ID:dvi_21909 |
Coordinate:scaffold_13052:767199-767349 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.8 | -29.2 | -29.2 |
 |
 |
 |
CDS [Dvir\GJ21440-cds]; exon [dvir_GLEANR_68:1]; intron [Dvir\GJ21440-in]
| Name | Class | Family | Strand |
| Helitron1_Dmoj | RC | Helitron | - |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTCAATTACTCAGAGCTTAAAATATGGAAGTGCAAATGATTCATTTATAAGTAAGTATACGTACACCCAAATGTATGTATATGGTATGTGCGCATTTTTAGGAGGGCTCAAAGAACAATATAGATCAATTTAAAAGTTCTGTTTTCTAGTTTGTCAACAAAGTAAAAAAAATAATTGGAGTTGTGCAATATCCGCTTTCTTGTCCCTTAGTCTGTCCGTCTTCGTTCCTTCCATCCTTGCGAATGAAAAAA **************************************************....(((((((((((......)))))))))))(((((.((((((.((((((((((((....(((((...................)))))))))))((.((((....)))).))..........)))))).))))))))))).........************************************************** |
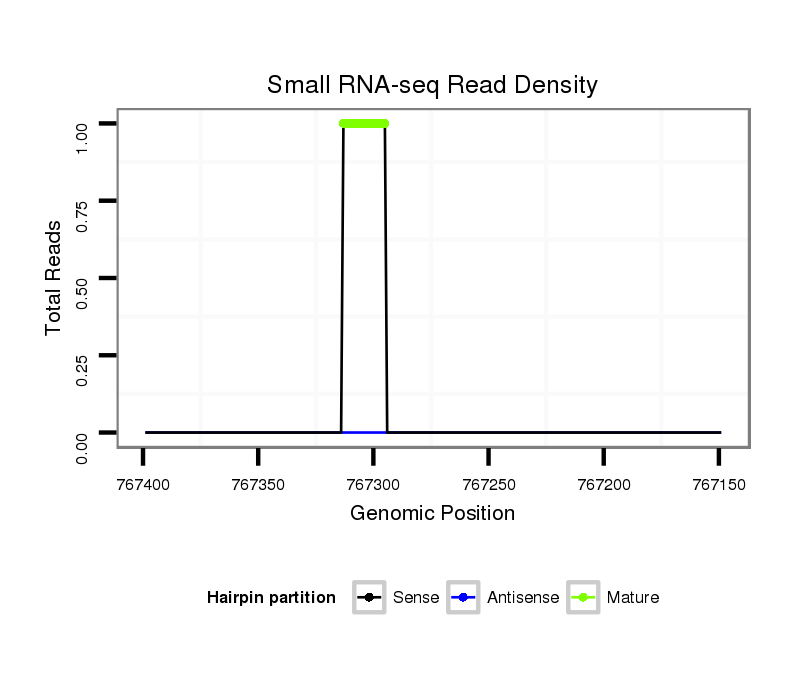
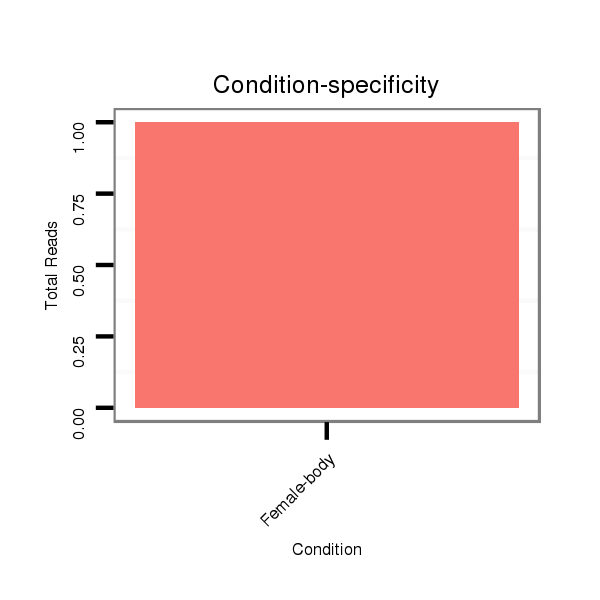
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060664 9_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
SRR060666 160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060657 140_testes_total |
SRR060674 9x140_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
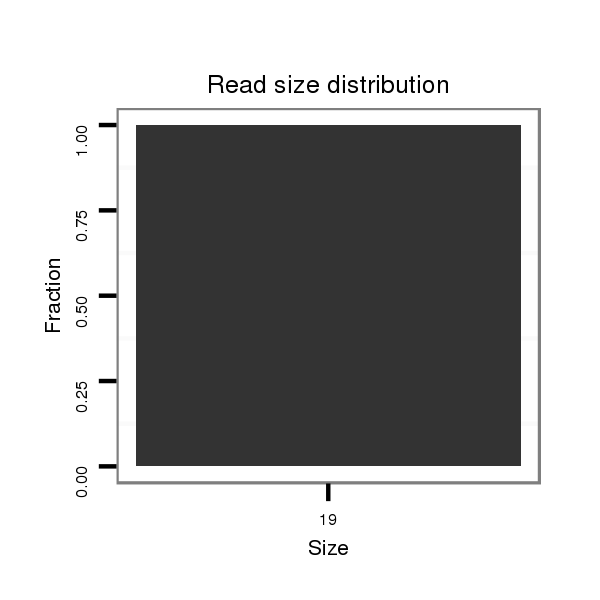
| ......................................................................................TGTGCGCATTTTTAGGAGG.................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CTCAGAGCGGAGAATATGGA............................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.50 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CTATCTGGCTTCGTTCCTTC.................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CTCAGAGCGGAAAATTTGGA............................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.18 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GTCTGTCCTTCTTCGTTTC....................... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............GCTTAAGAGATCGAAGTGCA......................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................TTCCATCCTTGGGATTGTAAA.. | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........CTCAGAGCTGAGAATTTGGA............................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CTCAGAGCGGAGAATATGG................................................................................................................................................................................................................................ | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................TATGTCAACAAAGGAAAA.................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................................................................................................ATCCTTGGGAATGAAGATA | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GAGTTAATGAGTCTCGAATTTTATACCTTCACGTTTACTAAGTAAATATTCATTCATATGCATGTGGGTTTACATACATATACCATACACGCGTAAAAATCCTCCCGAGTTTCTTGTTATATCTAGTTAAATTTTCAAGACAAAAGATCAAACAGTTGTTTCATTTTTTTTATTAACCTCAACACGTTATAGGCGAAAGAACAGGGAATCAGACAGGCAGAAGCAAGGAAGGTAGGAACGCTTACTTTTTT
**************************************************....(((((((((((......)))))))))))(((((.((((((.((((((((((((....(((((...................)))))))))))((.((((....)))).))..........)))))).))))))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060689 160x9_testes_total |
SRR060664 9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................CTGAACACGTTATAGGCGGA...................................................... | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .....................................................................................................CTCCCGGGTTTCTAGCTATAT................................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ..............................................................................................AAAAATCCGCCCGAGGTT........................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 |
| .........................................................ATGCATGTGCGTGTACATA............................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13052:767149-767399 - | dvi_21909 | CTCAATTACTCAGAGCTTAAAATATG------------------------------GAAGTGCAAATGATTCATTTA---------------TAAGTAAGTATACGTACACCC-AAATGTATGTATATGGTATGTGCGCAT---------TTTTAGGAGGGCTCAAAGAACAA-TATAGATCAATTTAAAAGTTCTGTTTTC-TAGTTTGTCAACAAAGTAAAAAAAATAATTGGAGTTGTGCAATATCCGCTTTCTTGTCCCTTAGTCTGTCCGTCT-TCGTTCCTTCCATCCTTGCGAATGAAAAAA |
| droMoj3 | scaffold_6496:26618808-26619070 - | TCGAGAAA----------------TA------------------------------TAAATATAAATAATTTACACAAATTTATGTTTGAGACAAACAAATACCTGAACATTCGAGATATTTAGCTTTTGTACGTACGCGCACAATTATTATTTATAAAGTTTAAAAGTCCAAAAATATAAAAAGCCAAAAATACACTTTTTATACTCATTCACGAAAGTGAATCAGTATATTGGTTTTGTGCAATGTCCGTCTTGCTGTCCGTCCGTCTGTCCGTCTGTCCGTCCGTCCGTCTGTCCGTATGAACGCA | |
| droGri2 | scaffold_14822:175720-175776 + | ATCCACTTCGCAAAACTATAAAAACG------------------------------GGAATGTAAATGAGACATATC---------------TAAGTAAGTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004943:1184738-1184802 + | TACAAGTGCCCAATGCGCAA--AATG------------------------------GCAATGTGAATAAATCAAGCT---------------TGAGTAAGTACACATATATC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_42:99448-99512 + | TTTAATCACCCTAAACGACAAACATG------------------------------GAAATTTGAATCATACAAATC---------------TAAGTAAGTATTTATATA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779182:64809-64865 + | CAGGATTCCATATTAACACATTAATA------------------------------GAA-----AAATGTCCGGATA---------------TAAGTAAGTATACAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | 4:224377-224436 + | CATTTTAACACATTACCAGATTCCAG------------------------TAAGTATAAGTGCATATTGTTT---------------------AATT-------CATTTATA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4512:1023561-1023641 - | CATAGCAATGC--------------TGGAGAGCAATTTAACACATTTACA------AAC-----AAATTCCTGGATT---------------CGAGTAAGTATATATGTA------------------------TGCATAT---------TTTT----------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:59 AM