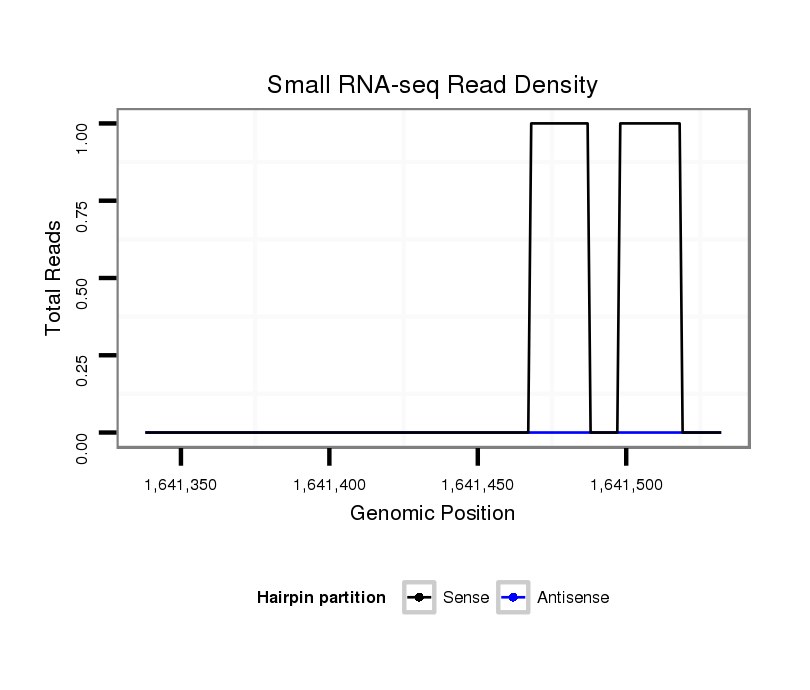
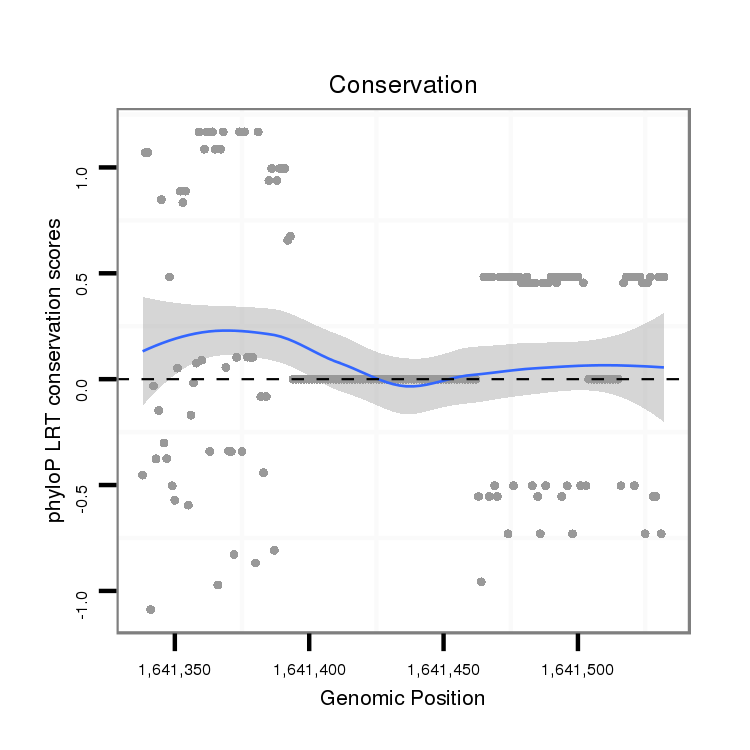
ID:dvi_21902 |
Coordinate:scaffold_13052:1641388-1641482 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_165:1]; CDS [Dvir\GJ16077-cds]; CDS [Dvir\GJ16077-cds]; exon [dvir_GLEANR_165:2]; intron [Dvir\GJ16077-in]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------------------------------################################################## TTACTTCCAATGCCAGTGCCGACGAGTCGGATACTGACAATGTTAGTCATGTAAGTGATATCCAAAATTTTTGAAACGCTTCACATAAATAGTTAAATTGTTGTTATGTATATAATTCGGTTGTGACTAATTTTTTGTGTTGCAGTCTTCCCATCATACTTTAGCAACGGCGTCACCTACAAAGAAGCGTTATAA **************************************************...........................((..((((.((((((((((((((..(((((....))))).))))).))))))..))).))))..))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060665 9_females_carcasses_total |
SRR060688 160_ovaries_total |
V053 head |
SRR060672 9x160_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060689 160x9_testes_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................TTTTTTGTGTTGCAGTCTTC............................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TTAGCAACGGCGTCACCTACA.............. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................ATACAAAATTTTTGAAACGCTT.................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................GCTAGTCAAGTAAGTGAT........................................................................................................................................ | 18 | 2 | 5 | 0.40 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................TTTTGCGTTGCAGTCCTC............................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................AGCAACGCCGTTACCTTCA.............. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................CGCTAGTCAAGTAAGTGAT........................................................................................................................................ | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................ACAATTCGGTTCTGACT................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AATGAAGGTTACGGTCACGGCTGCTCAGCCTATGACTGTTACAATCAGTACATTCACTATAGGTTTTAAAAACTTTGCGAAGTGTATTTATCAATTTAACAACAATACATATATTAAGCCAACACTGATTAAAAAACACAACGTCAGAAGGGTAGTATGAAATCGTTGCCGCAGTGGATGTTTCTTCGCAATATT
**************************************************...........................((..((((.((((((((((((((..(((((....))))).))))).))))))..))).))))..))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060689 160x9_testes_total |
SRR1106716 embryo_4-6h |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................ACAACAGAACGTCAGAAGGGAA......................................... | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................AAATTTTGCGATGTGTATT........................................................................................................... | 19 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................TCCGCATTGGATGTTTC........... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................TCAGACCGGCAGTATGAAA................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13052:1641338-1641532 + | dvi_21902 | TTACTTCCAATGCCAGTGCCGACGAGTCGGATACTGACAAT---GTTAGTCATGTAAGTGATATCCAAAATTTTTGAAACGCTTCACATAAATAGTTAAATTGTTGTTATGTATATAATTCGGTTGTGACTAATTTTTTGTGTTGCAGTCTTCCCATCATACTTTAGCAACGGCGTCACCTACAAAGAAGCGTTATAA |
| droMoj3 | scaffold_6498:696661-696732 + | ATAATATCGTTATT------------------------------------------------------------------------------------------------------------------TGTATTCATTTTTATTGCAGCCAGCTCATCAAATTGTAACG------------GCAAAAAAGAGTATTCA | |
| droGri2 | scaffold_14822:657046-657081 + | TTAG--------CCAGTTTCAATGAATCCGATGTCGATAAT---GCT------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004943:1754236-1754280 - | TTAGCTCCAA---------CAACGAATCGGAGACGGACAAT---GCTGCCCAGGTAA--------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | Unknown_group_43:60039-60086 + | G-----------ACAGTTCGAACGAGTCAGATGTTAATAGAAGGAGTACTCACGTAAGT------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 05/16/2015 at 09:27 PM