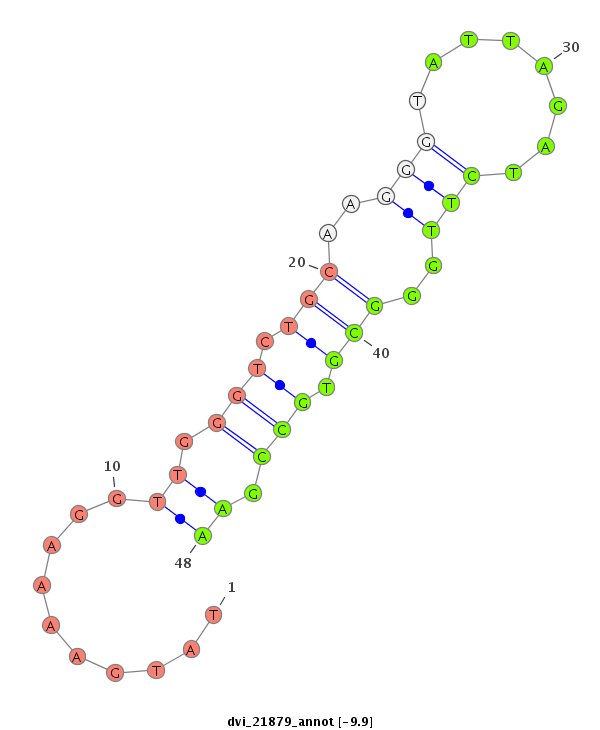
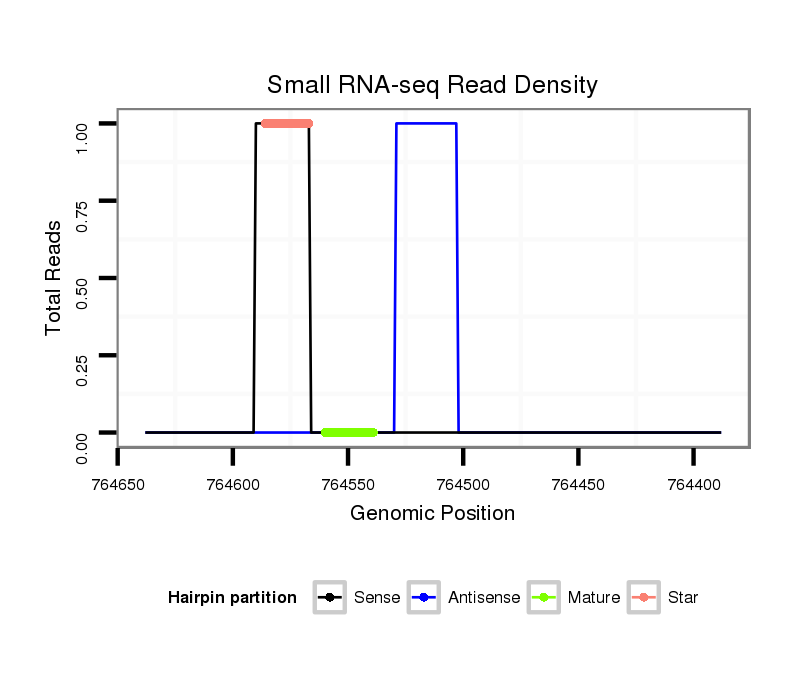
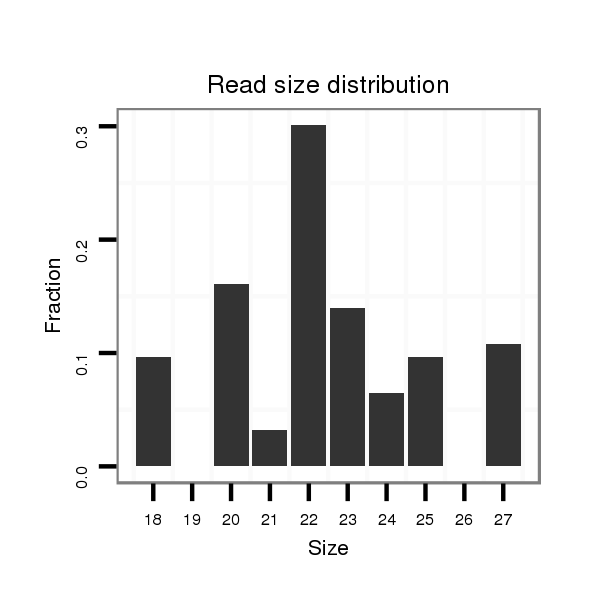

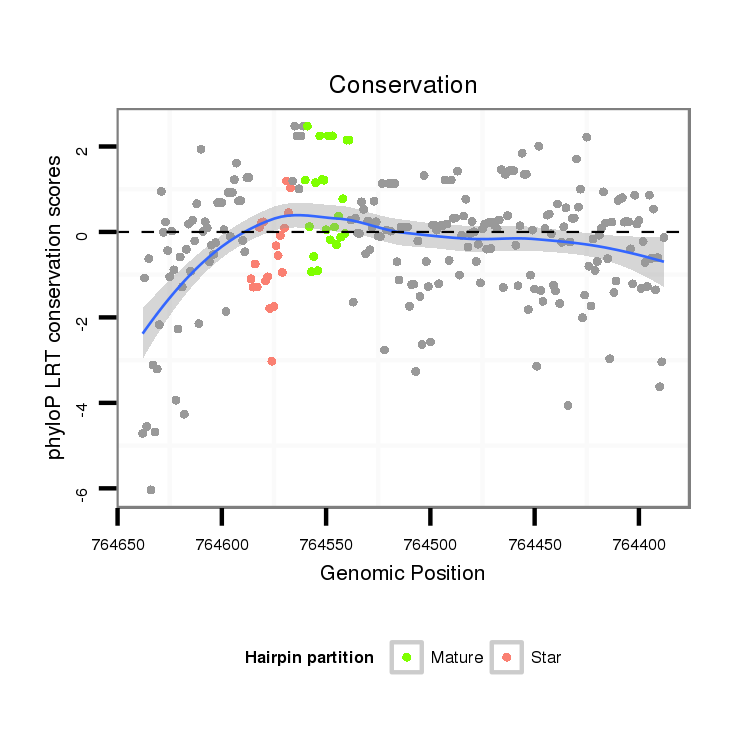
ID:dvi_21879 |
Coordinate:scaffold_13052:764438-764588 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.4 | -9.1 |
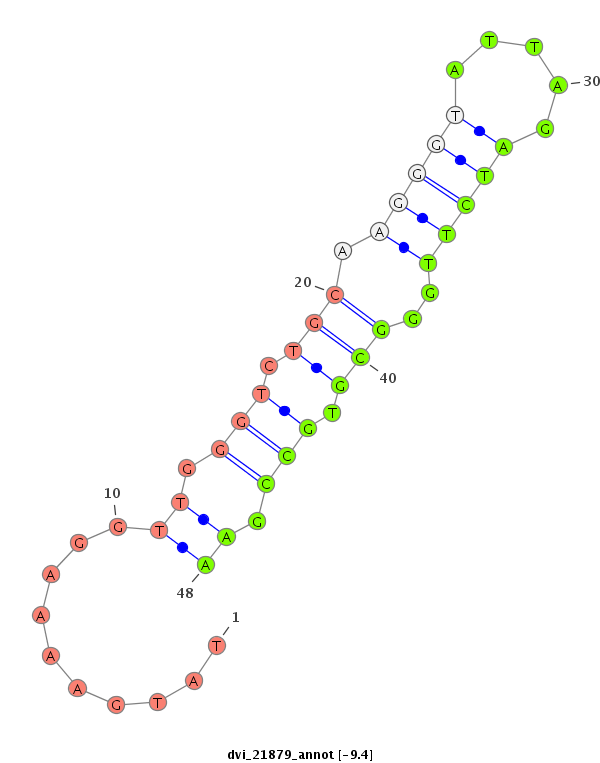 |
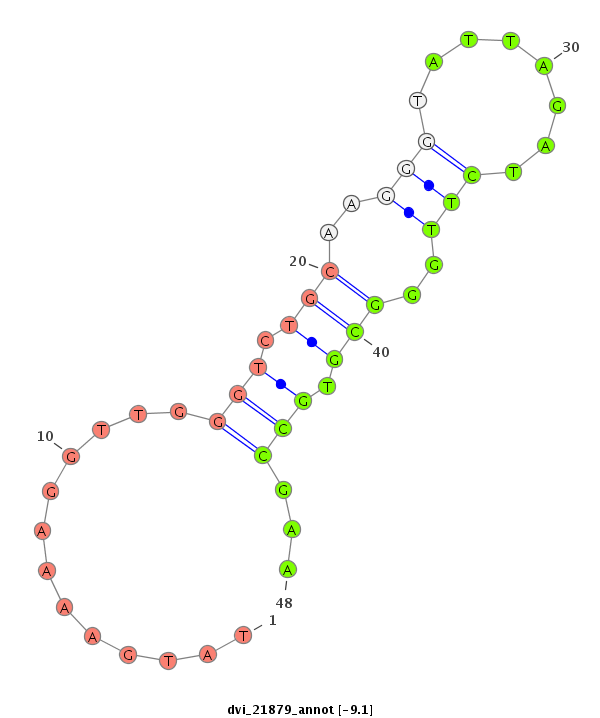 |
exon [dvir_GLEANR_68:2]; CDS [Dvir\GJ21440-cds]; intron [Dvir\GJ21440-in]
| Name | Class | Family | Strand |
| Helitron-2N1_DVir | RC | Helitron | - |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CATTTACTATTTTCCCGGTACTTCTTAGATAGGGGCAAAGCACTATGAGCATTATGAAAAGGTTGGGTCTGCAAGGGTATTAGATCTTGGGCGTGCCGAAGATAGCCCTTCTTTCTCGTTTTCATTTTGTTGTGGTTTTTCAGATTAATTTACATATATAATATACTGAGCTTTTTTTTTCATTTTGTTGTGATTTTTCAGATTCTAAAAATGAACCGACAAATATGCAAAAATCCAAATCGTTATCTAAT ****************************************************..........((.(((.(((..(((........)))..))).))).))******************************************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060674 9x140_ovaries_total |
M047 female body |
SRR060679 140x9_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060678 9x140_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060670 9_testes_total |
SRR060658 140_ovaries_total |
SRR060675 140x9_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
V116 male body |
M027 male body |
SRR060669 160x9_females_carcasses_total |
SRR060655 9x160_testes_total |
SRR060657 140_testes_total |
SRR060673 9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................GCATTATGAAAAGGTTGGGTCTGC................................................................................................................................................................................... | 24 | 0 | 20 | 1.25 | 25 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TGAGCATTATGAAAAGGTTGGGTC...................................................................................................................................................................................... | 24 | 0 | 20 | 0.35 | 7 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TGAGCATTATGAAAAGGTTGGGTCT..................................................................................................................................................................................... | 25 | 0 | 20 | 0.30 | 6 | 0 | 0 | 0 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................ATTAGATCTTGGGCGTGCCGAA....................................................................................................................................................... | 22 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCATTATGAAAAGGTTGGGTCTGC................................................................................................................................................................................... | 25 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGCAAGGGTATTAGATCTTGGGCGTGC........................................................................................................................................................... | 27 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TCTGCAAGGGTATTAGATCTTG.................................................................................................................................................................. | 22 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGCAAGGGTATTAGATCTTGGGC............................................................................................................................................................... | 23 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TATGAAAAGGTTGGGTCTGC................................................................................................................................................................................... | 20 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCTTAGATAGGGGCAAAGCACTAT............................................................................................................................................................................................................. | 24 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TGAAAAGGTTGGGTCTGC................................................................................................................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCATTATGAAAAGGTTGG......................................................................................................................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TTTACTATTTTCCCGGTACTTCTTAGA.............................................................................................................................................................................................................................. | 27 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGCAAGGGTATTAGATCTTGGGCC.............................................................................................................................................................. | 24 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................ATAGATAATATACGGATCTT.............................................................................. | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................GGCAAAGCACTATGAGCA........................................................................................................................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............TCCCGGTACTTCTTAGATAGGGGCA...................................................................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................ATGAGCATTATGAAAAGGTTGGG........................................................................................................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TAGGGGCAAAGCACTATGA........................................................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CTTAGATAGGGGCAAAGCACTATG............................................................................................................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................TTCTTAGATAGGGGCAAAGCACTATGA........................................................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TTTTCCCGGTACTTCTTAGATAGG.......................................................................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .ATTTACTATTTTCCCGGTACTTCTTAGA.............................................................................................................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TAGATAGGGGCAAAGCACTATGAGCAT....................................................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GTTGGGTCTGCAAGGGTATTAGATC..................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGGGTCTGCAAGGGTATTAGA....................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................ATAGGGGCAAAGCACTATGAG.......................................................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGGGTCTGCAAGGGTATTAGATCTT................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTATGAAAAGGTTGGGTCTG.................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................GCACTATGAGCATTATGAAAAGGTTGGG........................................................................................................................................................................................ | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TGAAAAGGTTGGGTCTGCAA................................................................................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATTATGAAAAGGTTGGGTCTGC................................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................TACTTCTTAGATAGGGGCAAAGCAC................................................................................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TATGAAAAGGTTGGGTCT..................................................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATTATGAAAAGGTTGGGTCTGCA.................................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AGCACTATGAGCATTATGAAAAGGT............................................................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCTTAGATAGGGTCAAAGCACTATGAGC......................................................................................................................................................................................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CATTATGAAAAGGTTGGGTCTGCA.................................................................................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TGAAAAGGTTGGGTCTGCAAGGGTA............................................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAAGGTTGGGTCTGCAAGGGTATT.......................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TATGAGCATTATGAAAAGGTTGGG........................................................................................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................TTGGGTCTGCAAGGGTATTAGATC..................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TGAGCATTATGAATAGGTTGG......................................................................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TTTACTATTTTCCCGGTACTTGTTAGA.............................................................................................................................................................................................................................. | 27 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GTAAATGATAAAAGGGCCATGAAGAATCTATCCCCGTTTCGTGATACTCGTAATACTTTTCCAACCCAGACGTTCCCATAATCTAGAACCCGCACGGCTTCTATCGGGAAGAAAGAGCAAAAGTAAAACAACACCAAAAAGTCTAATTAAATGTATATATTATATGACTCGAAAAAAAAAGTAAAACAACACTAAAAAGTCTAAGATTTTTACTTGGCTGTTTATACGTTTTTAGGTTTAGCAATAGATTA
*******************************************************************************************************************************************************..........((.(((.(((..(((........)))..))).))).))**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060677 Argx9_ovaries_total |
M047 female body |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
SRR060675 140x9_ovaries_total |
SRR060676 9xArg_ovaries_total |
SRR060670 9_testes_total |
SRR060678 9x140_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060656 9x160_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060655 9x160_testes_total |
V053 head |
SRR060673 9_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060683 160_testes_total |
SRR060654 160x9_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060669 160x9_females_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060688 160_ovaries_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................AGAAAGAGCAAAAGTAAAACAACACCA................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TACTTTTCCAACCCAGACGTTCCCAT............................................................................................................................................................................ | 26 | 0 | 20 | 0.35 | 7 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CCCGTTTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 26 | 0 | 20 | 0.30 | 6 | 0 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CCCCGTTTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 27 | 0 | 20 | 0.25 | 5 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................GCGAAAAATAAAGTAAAACAAC............................................................. | 22 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CCCGTTTCGTGATACTCGTAA...................................................................................................................................................................................................... | 21 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................TCGTGATACTCGTAATACTTTTCCAA........................................................................................................................................................................................... | 26 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................GTTTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 23 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CCGTTTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 25 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................CGGCTTCTATCGGGAAGAAAGAGCAAAA................................................................................................................................. | 28 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................CTTCTATCGGGAAGAAAGAGCAAA.................................................................................................................................. | 24 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGAAAGAGGAAAAGTAAAAC.......................................................................................................................... | 20 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................ACTCGTAATACTTTTCCAA........................................................................................................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CGTTTCGTGATACTCGTAATACTTTT............................................................................................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................GCTTCTATCGGGAAGAAAGAGCAAAA................................................................................................................................. | 26 | 0 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AATGATAAAAGGGCCATGAAGAATCT.............................................................................................................................................................................................................................. | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GGCTTCTATCGGGAAGAAAGAGCAAAA................................................................................................................................. | 27 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CGTTTCGTGATACTCGTAATACTTT................................................................................................................................................................................................ | 25 | 0 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TTCGTGATACTCGTAATACTTTTCC............................................................................................................................................................................................. | 25 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGTAATACTTTTCCAACCCAGACG................................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CGTTTCGTGATACTCGTAATACTTTTC.............................................................................................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CCGTTTCGTGATACTCGTAATACTTT................................................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GTAGAATCTATCCCCGTTTCGTGATACT........................................................................................................................................................................................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GGGCCATGAAGAATCTAT............................................................................................................................................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTTCGTGATACTCGTAATACTTTTCC............................................................................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AAGAGCCAAAGTAATAC.......................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................CCCGTTTCGTGATACTCGTAAT..................................................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................CGTGATACTCGTAATACTT................................................................................................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........AAGGGCCATGAAGAATCTATCC.......................................................................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTTCGTGATACTCGTAATACTTTTCCAA........................................................................................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CCGCACGGCTTCTATCGGGAAGAAA......................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GCACGGCTTCTATCGGGAAGAAAGAG...................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CCCGTTTCGTGATGCTCGTAATA.................................................................................................................................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CCGTTTCGTGATACTCGTAA...................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TTCCAACCCAGACGTTCCCATAATCTAGA.................................................................................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CCCGTTTCGTGATACTCGTA....................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TCCCCGTTTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TACTTTTCCAACCCAGACGTTCCCATA........................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......ATAAAAGGGCCATGAAGAAT................................................................................................................................................................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................ACTTTTCCAACCCAGACGTTCCCAT............................................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........TAAAAGGGCCATGAAGAATA............................................................................................................................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TCTATCGGGAAGAAAGAG...................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTATCGGGAAGAAAGAGCAAAA................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GGCTTCTATCGGGAAGAAAGAGCA.................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................CGGCTTCTATCGGGAAGAAAGAGC..................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................CACGGCTTCTATCGGGAAGAAAGAGC..................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................TCGTGATACTCGTAATACTTTTC.............................................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................ATCCCCGTTTCGTGATACTCGTAATACTT................................................................................................................................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CGTTTCGTGATACTCGTAATA.................................................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GAAGAATCTATCCCCGTTTCGTGATACT........................................................................................................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AATGATAAAAGGGCCATGAAGAAT................................................................................................................................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................GTTTCGTGATACTCGTAATA.................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CTGATAAAAGGGCCATGAAGAATCT.............................................................................................................................................................................................................................. | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13052:764388-764638 - | dvi_21879 | CATTTACTATTTTCC--CGGTACTTCTTA-------------GATA-------------------------GGGGCAAAGC--ACTA----TGAG-CAT----------TATGAAAAGGTT-GGGTCTGCAAGGGTA----TTAG-ATCTTGGGCGTGC-CGAAGATAGCCCTTCTTTCTCGTTTTCATTTTGT--------------------------------------TGTGGTTTTT--------------------------------------------------------------------------------------------------------------C-AGATTAATTTA---CAT--ATAT-------------------------------------------------AATATACTGAGCT----------------------------------------------------------------------------------------------------------------TTT-TTTTTCATTT--TGT-TGTGATTT-----------------------------------------------------------TTCAGATT---C--TAAAAATGAAC-C---GACAAA--------------------------------------------TAT--GCAAAAATCCAAATCGTTATCTAAT |
| droMoj3 | scaffold_6498:1444791-1444932 + | TTTTGTTTT--TGCC--TT--GTCCTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACGTTGATAAATCTACCATTCATTGATATATCCCTAATTGATCCTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT--TTTTCGGTTCACTT-----------------------------------------------------------T-CAGATTCAGA--TAAAAATGAAA-C---GACAAA--------------------------------------------TAT--GCAGAAATCAAAATCTTTATCTAAT | |
| droGri2 | scaffold_15112:3689080-3689300 + | TTTCACGGC------------AATT----------------TAAC--------------------------------------------------------------------------AG-CGATCTACAAGGGTA----TTTA-AGCTTCGGCGTGT-CTAAGACTG----------------------------------------------------------------------------------------------------------------GCATTCTGTAC-TTGTTT-ATG-G--CA---------------------------------------------------------------------------------------------------------------------------ACATTGTTTGTTTGTTTTTCTATTACGCAATATTCGCAT-------------GGAATG----------CTCGA--------------------------------------------------------AT--TTG-------TCAAACTTGAT----------------------------------------T-------ATTACATACTTG---TTAAGAAAAGGAAA-C---AACAAA-AATGAAA----------------------ACCAA---------AAC----CAAAACCAAAACAACTATCTACT | |
| droWil2 | scf2_1100000004953:1067207-1067464 - | CCTTTTAGC--TTAA--AC--GTTTTTC--------------CA---CTTT---------GA---AGGCTATAGGTAAAGG-----AGAACT----------------TACCAA------AA-AAGTTGCAAGGGTA----TACA-AACTTTGACGCGGTCGAAGTTAGCC-CCGGCCC----------------------------------------------------------------------------------------------------------------TCTGGTTT-TT-----AGTTTATTTTGATTTTTTTTGTAATTTTCTTAATCTA-----------------------------ATATGAAA----------TTGTG--------------------------------------------------------------CCAATACTTTTTTGGCGTGAACATGTGC---CT-------ATT-----------------------------------TT-------------------------------------------------------------------------------TTAGT----------------------TT---A--TAG--AACATAAC---TGCAAA-------------------------AAAATGCTAGGAAAATGAATT---------------------TTTATTT | |
| dp5 | XR_group6:10778722-10778837 - | CCCATTTTT--TTTA--TTTTCCTTTTTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGTTTTTTCATTC----------------------------------------------TTTT-------------------------------TTTTAT--------------------AT-----------------ATAT-------------------------------------------------AT-------------------------------------------------------------------------------------------------------------------------------TTTTTAAT--TTG-------ATTTGTTTGAT----------------------------------------T----------------------T--T------------ATT--T--GTTAATTAATTTTCCTCAAATTCCTA----------------------------------------------AGA | |
| droPer2 | scaffold_3:215264-215446 + | ATAAAAACT--------------------------------------------------------------------------------------------------------------CT-CGATCAGCTAGGGTA----TAAAAAGCTTTGCCGCGC-CGAAGCTGGCT-TCCTTATTTGTTTTTATTTTT------------------------------------GT-TGTTGTT---------------------------------------------------------------------------------------------------------------------------TAAAA-TGT--GC---AGCTTTTGGCAATTTTTCTTTAAAATTTTGTTCC--------------------------------------------------------TTT-------------------------------------------------------------------ATAGCGACGTTCTTGT--------TTT--TGT-TTTTGTTT-----------------------------------------------------------TTGTAATT---C--GAG---------------------------------------------------------------CT---------------------ATGACCC | |
| droAna3 | scaffold_13260:1938536-1938768 + | TAAGGATCG---GCC--GA----------------------------CTATATCCTATAGCTGCC--------------ACA-----TAACTGAA-CAT----------CCGGAATT-GGCA-TAACTGCAAGGGTA----TATC-AACTTCGGCTCC----------------------------------------------------------------------------------------------------------------GCCCAAAGTTAGCTTTCCTTTC-TTGTTT-TTTTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGTTTATAATATAGTAAAAGTCTCATAAGAATAGGCCTCTCATCTGTTTGAGACAATTTGGTTTCCCACAA------------CTAAAA-A---GACGGA--------------------------------------------TGTGTGCAAAGTTTCAACTCGATA---GCT | |
| droBip1 | scf7180000395032:5212-5451 + | TATATATTC--CTAT--TCCCAT----AAGGATCGGCC-------AACTATATCCGATGTTTGCG--------------ATAT------------ATAT----------C------CGGTT-TTAACTGCAAGAGTA----TATA-AACTTCGGCTCC----------------------------------------------------------------------------------------------------------------GCCCGAAGTTAGCTTTCCTTTC-TTGTTT-TTATT-------------------------------------GAA-----------------------------TGGG----------AAATTCTGATTATAAATTTGTCGGAT--------------------------------------------------------------------------------------------------------------------------TTAGAACAAGGACCATATATTTTACATTT--TTT-TATAATCTTTTG--------------------------------------------------------------------------AACATA--A--GCC-AA--------------------------------------------TAT--TTAAAATATCA-AA---TA-TCAAT | |
| droKik1 | scf7180000302441:526004-526241 + | TTTAGTGTC--TGTT--TT--CGGC------------A---------TTAT--------------AATTTATAAT----ATAA------------ATAT----------G------AAAAC-CAATCTGCAAGGGTA----TACA-AACTTCGGCGTGC-CGAAGTTAGCT-TCCTTTC------------------------------------------------------------------------------------------------------------------TTGTTT-TTTAA-------------------------------------TAAAGTTAATCAATATTTATAAAAA-TGT--AC---ACTATTAGGA--AATGT---------------------------AATTAACCATGTCTTCTTTTCATTA------------------AA--------------------------------------------------------------------------------------------------------AATGATTT-----------------------------------------------------------GGT-A------A--TAAAAAAAATG-C---ATCA-A--------------------------------------------AGT---AAAAAATTCAAATTATTTACTCAT | |
| droFic1 | scf7180000453473:6298-6527 - | AATATAGAG--TTTTCTAA--CGAC------------TCTTGAATAGCA----------------------------------AATAAAATT----------------TTCGAAATATTTGA-TGACTGCAAGGGTA----TATA-AGCTTCGGCTTGC-CGAAGCTAGCT-TCCTTTC------------------------------------------------------------------------------------------------------------------TTGTTT-TT-----CGCTTTTTTT--------------------------------------GAT------------------------------AAACTCTAATTTTAAACCTATTGAATTGAAACT-------------------------------------------------TGGTTTGAAT------------------AC---------------------------------------------------------------------------ATTT-----------------------------------------------------------TGTAAGCT---C--CCAAGATGAAC-T---ACCAAC--------------------------------------------TTT--CAAATAATTTGGGGCACTATTTCAT | |
| droEle1 | scf7180000491347:79955-80194 - | AAATCATCATAGCTT--CAATGTTTTTA--------------AA----------------------------------------------------CATATACGCAAGTAAATTCTT-GCAA-TAGCTGCAAGGGTA----TATG-AACTTCGGCTTGC-CGAAGTTTGCT-TTCTTTC------------------------------------------------------------------------------------------------------------------TTGTTT-TTTTT-------------------------------------AGAAGTTCTTCTAGACTTATTAATA------ATATGACAGTTGAGA--ATTACGATTTTAATTATATTGAAA-----------------------------------------------------------------------------TCGGATTAC--------------------------GATATCAGAATG-------------------------------------------------------------------------------CTGCC---------------------------A--T----AGGAAC-T---GACGAA--------------------------------------------TAT--TTAA-GCTGCAAATCA-----TCAT | |
| droRho1 | scf7180000776960:102198-102429 - | AACTTTGGA--TTTT--TTATAGGTTTTATTTTTGGCC-------AACT----------------------------------AATATAATT----------------TACAGA------A-TATTCTGCAAGGGTA----TA-----CTCCGGCTCC----------------------------------------------------------------------------------------------------------------GCCCGAAGTTAGCTTTCCTTTC-TTGTTT-ATTTT-------T-------------------------------------TTTTCGTTTTTTTAA---GGC--ACGTGAAAGTCAAAA--ACACCGATTT-----------------------------------------------------------------TCGCTC-------------AAAACG-TCGACTTATT-----------------------------------TG-------------------------------------------------------------------------------TTGCT---------------------------C--TAAAAATA---------ATAAA-----------------TTTTCAAAAAA-------------------------AGCTCGACAGCGTTACCT-GA | |
| droBia1 | scf7180000302098:48084-48306 + | TACCAACAA--TCCT--CAAAAATT-TCA-------------CATG--------------GTGCT--------------ACA----------------------AAAGTTGATTATTTCTTA-TAAGTGCAAGGGTT----TACC-AACTTCGGCTTGC-CGAATTTAACT-TCCTTTC------------------------------------------------------------------------------------------------------------------TTGTTT-TATTA--CA-----------------------------------A----------------------------------------------------------TCATCTCAAGCTGCAACTTAGGTAGCATTATATTCCTTCAATTT------------------TG--------------------------------------------------------------------------------------------------------ACTGAATA-----------------------------------------------------------TGCACAAT---T--TAAAAAAGATA-----TACAAA--------------------------------------------AAT--ATAAAAATATA-ATTAGTTTCT-AA | |
| droTak1 | scf7180000415288:115747-115986 - | AAATTAACA--TTTT--TGCGATTTG-TA-------------AA------------------------T--------------------------T------------AAT-----AGGAA-TTATCTGCAAGGGTA----TATA-AGCTTCGGCTGGC-CGAAGCTAGCT-TCCTTTC------------------------------------------------------------------------------------------------------------------TTGTTT-TATTT--CACTTTTTTT------CCTTGAAACC--ATTAAT-------------------------ATTATCGATACGATA----------TTGT-----------------------------------------------------------------------------------AGGTATGG---AT-------TTTTTTAGTTTTACTTATTGATAGTCATG-------ATCTCTACCAATGACCAT--------------------------------------------------------------------------------------------------------------GAC---TATCAATAAGTAAA----------------------ACTAAAAAAATCCATA--------------------CCTACAAT | |
| droEug1 | scf7180000408693:30-118 + | TTTTTTATA--TTTT--TAATATTTTTTAT-------------A---------------TTTTTT--------------ATA-----TTTTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATATTTT--------------------TT-----------------ATAT-------------------------------------------------TT-------------------------------------------------------------------------------------------------------------------------------TTTTATAT--TTTTT-TATATTTT-----------------------------------------------------------T-------------------------------------------------------------------------------TT---------------------ATATTTT | |
| dm3 | chr3L:18848928-18849078 - | TATTCGTTA--ATAT--TGGAATTTAATGT-------------A--------------------------------------------------------------------------TAAA-TAACTGCAAGGGTACGTATGAA-AGCTTCGGCATGC-CAAAT-TATCT-TACTTCC-------------------------------------------------------------------------------------------------------------------TGTTTATATTTTTTA----------------TT-TTTTAT--------------------AT-----------------ATTT-------------------------------------------------AATATTATATTCTTTCTTAAT-----------------------------------------------------------------------------------------------------------------TTTTT--AAT-TATGATT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------TTT | |
| droSim2 | 2r:5310639-5310794 - | AAGTGATTGTATTAT--TAATAGTTTGTA-------------CA--------------------------------------------------------------------TA----ATA-TGTAGTGCAAGGGTA----TTTA-AGCCTCGGCTTGG-CAAAGCTAGAT-ATTTTTCTGGTTTTCAAAAATT--------------------------------------TTAAATTTTA--------------------------------------------------------------------------------------------------------------C-AA--TTTTTAA---GGT--ACAT-------------------------------------------------AA---------------------------------------------TATGTAA----A--GAG-T------AATTTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT---------------------TTTATTT | |
| droSec2 | scaffold_6:470009-470207 - | C------------------------------------------------------------------------------------------------------------------------------TTCAAGGGTA----TATA-AGCTTCGACTTGC-CGAAGCTAGCT-TTCTTTTTCGTTTTCGTTTTAAGTACACAGTATTTAAAATTTGTTGTGTTAACATTTAAAGCAAATTAAATAAATATACATTTACAAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCG-T------CAATCT-----------------------------------------------TTT-A-----AT--TTTTAAACTGGTTT-----------------------------------------------------------TTAATGA----C--CAAAAAACACG-T---GCCAAG--------------------------------------------TGC--TAAAAAATTTAAA------------ | |
| droYak3 | 2R:18125134-18125303 - | CATTGACTG--TTAT--TTTTCGGTA---------------------CAAT--------------TTTTTTTCTC----GAAC------------A------------TAC-----ATATA-TTAACTGAAAGGGTA----TATT-TGCTTCGACTTGC-GAAAGGCACCTGTTCTCTGTTTTATTTGTTTTAT--------------------------------------TTCTCTCTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCAAAATCTAGTTGATATGTACATATGT---AC-------ACTCTCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT---------------------TTCACTT | |
| droEre2 | scaffold_4770:16384575-16384702 + | TTATTTTTA--TTTT------ATTTTTTATTTTA--------TA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATATTAT--------------------AT-----------------ATAT-------------------------------------------------TATTTTATAT--T----------------------------------------------------------------------------------------------------------------TAT-TTTTTAGTTT--TTT-TGTAATTTTTATGAT----------------------------------------G----------------------T--T------------ATG--T--ATTGTAGAAATTAAAAAAAATTCAAA-------------------AA--------------------TGTCCAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 05:19 AM