
ID:dvi_21834 |
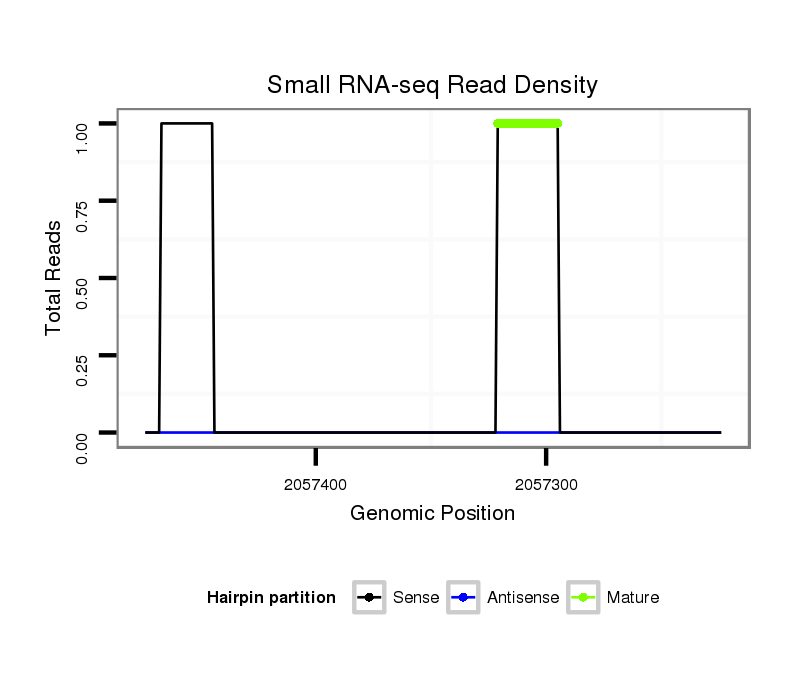
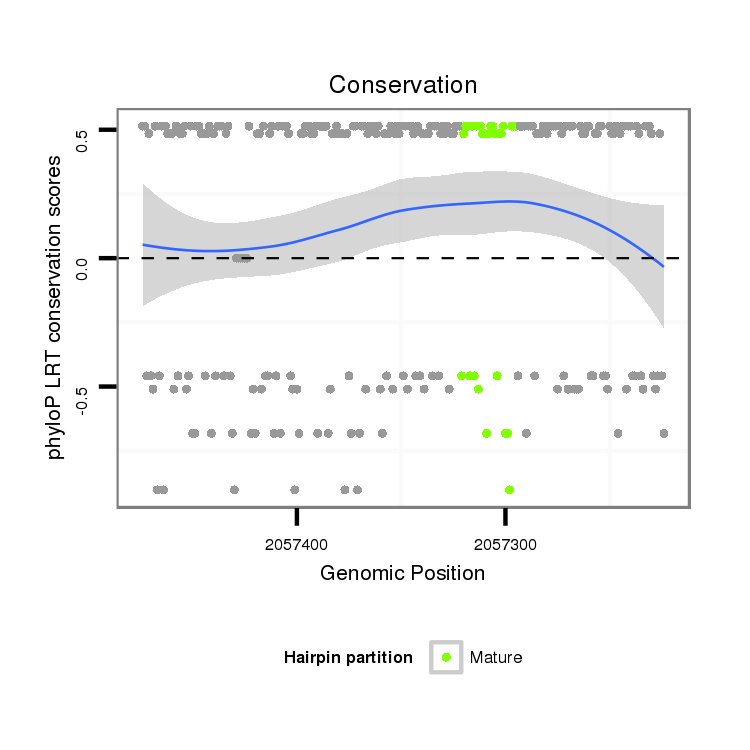
Coordinate:scaffold_13246:2057274-2057424 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.7 | -27.6 | -27.6 |
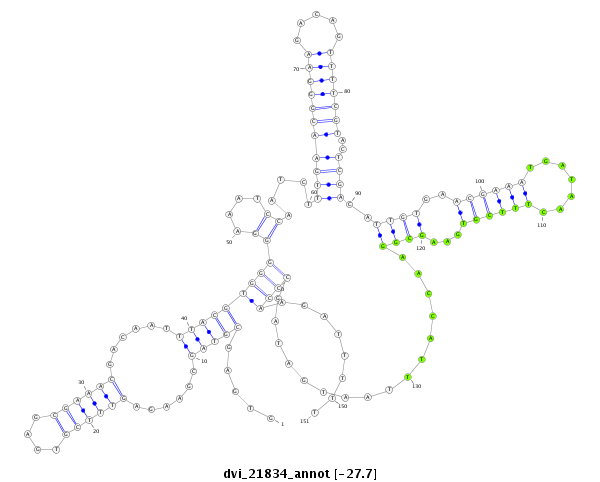 |
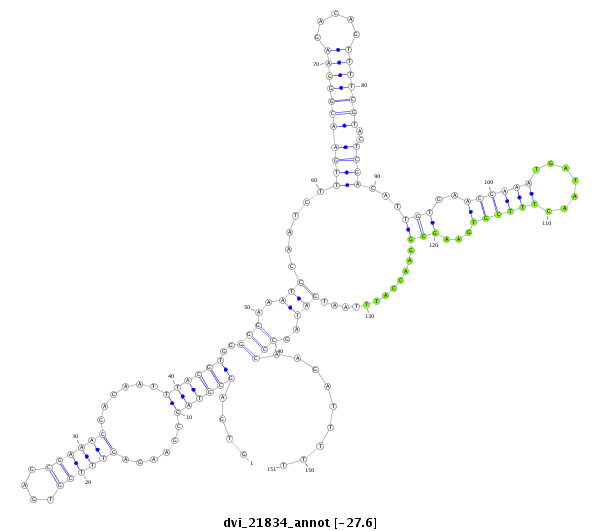 |
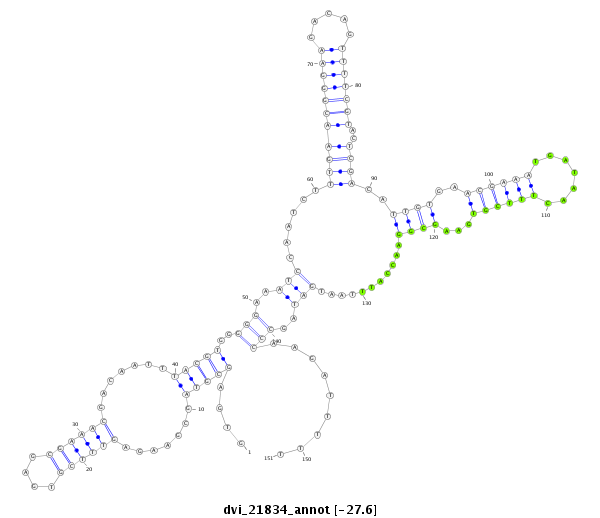 |
exon [dvir_GLEANR_16593:3]; CDS [Dvir\GJ16144-cds]; intron [Dvir\GJ16144-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAGCCATCGTGAGCGTAGCGATGAGTTTCGTGAGCAAAACGACAGATATCGTGAGCGTAGCGAAGAGTTTCGTGAGCGAAACGACAATTTACGTGGGGGAAATCCAATCTTTGAACGGGAAGACAGTTTTCGTACTCGACATTGTGAACGAAATGATAACTTTCGTGAAGCGGAACCATTTAATGATAGCCCAAGATTTTTGAAAAAATTTCGTAGCCCAAATTTGGACGATTTTGGTCCAAATCAATGCA **************************************************....((((((......((((((....))))))......))))))..(((..(((......(((((((((((.....)))))))..))))..((((..((((((.......))))))...))))...........)))..))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060661 160x9_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
M047 female body |
M027 male body |
V053 head |
SRR1106730 embryo_16-30h |
SRR060662 9x160_0-2h_embryos_total |
V047 embryo |
SRR060667 160_females_carcasses_total |
SRR060659 Argentina_testes_total |
M028 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......CGTGAGCGTAGCGATGAGTTTCG............................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AATTTGGACGAACTTGGTC............ | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TGATAACTTTCGTGAAGCGGAACCATT....................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GAATTTGGACGAACTTGGTC............ | 20 | 3 | 5 | 0.80 | 4 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GAACGGAAAGACAGTTTT......................................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................TTTGGTCCACATCAATGTA | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................GTGAAATCGAATCTTTGAACGC..................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GAGCGAAGCGGTGAGTTT............................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AATTTGGACGAACTTGGT............. | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................ATCGAATCTTTGAACGCGCA.................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................GTAGCGAAGGGTTTCGTGT................................................................................................................................................................................ | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................CATCGAATCTTTGAACGCG.................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................AATTTGGACAAATTTGGTC............ | 19 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................GATGGGTTTCGTTACCAAA..................................................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....CAAAGTGACCGTAGCGATG.................................................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CCCAAGATCATTGAAAAAATC......................................... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTCGGTAGCACTCGCATCGCTACTCAAAGCACTCGTTTTGCTGTCTATAGCACTCGCATCGCTTCTCAAAGCACTCGCTTTGCTGTTAAATGCACCCCCTTTAGGTTAGAAACTTGCCCTTCTGTCAAAAGCATGAGCTGTAACACTTGCTTTACTATTGAAAGCACTTCGCCTTGGTAAATTACTATCGGGTTCTAAAAACTTTTTTAAAGCATCGGGTTTAAACCTGCTAAAACCAGGTTTAGTTACGT
**************************************************....((((((......((((((....))))))......))))))..(((..(((......(((((((((((.....)))))))..))))..((((..((((((.......))))))...))))...........)))..))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060682 9x140_0-2h_embryos_total |
M028 head |
M047 female body |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
V116 male body |
SRR060666 160_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
V053 head |
M061 embryo |
SRR060657 140_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060678 9x140_testes_total |
SRR060689 160x9_testes_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................CCCTTCTGTCCTAAGCTTG.................................................................................................................... | 19 | 3 | 14 | 13.86 | 194 | 130 | 12 | 17 | 0 | 0 | 0 | 0 | 5 | 3 | 7 | 2 | 6 | 2 | 3 | 2 | 0 | 1 | 1 | 1 | 1 | 1 |
| ...................................................................................................................GCCCTTCTGTCCTAAGCTTG.................................................................................................................... | 20 | 3 | 3 | 3.33 | 10 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TCGCTATGCTGTTAAAAGCACC........................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................CGCTATGCTGTTAAAAGCAC............................................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTATGCTGTTAAAAGCACCC.......................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTATGCTGTTAAAAGCACCCAC........................................................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................CGCTATGCTGTTAAAAGCACCC.......................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................TTGCCGTTCTGCCAAAAGC....................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TCGCATTGCTGTTAATTACA............................................................................................................................................................. | 20 | 3 | 12 | 0.17 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TCGCTATGCTGTTAAAAG............................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAGAAGCATAAGCTGTCAC........................................................................................................... | 19 | 3 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GCCCTTCTGTCCTAAGCTT..................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACTAGCCCTTCTGTCCTAA......................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TGCTGTTAAAAGCACCCACA....................................................................................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TCGCATTGCTGTTAATTAC.............................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13246:2057224-2057474 - | dvi_21834 | AAGCCATCGTGAGCGTAGCGATGAGTTTCGTGAGCAAAACGACAGATATCGTGAGCGTAGCGAAGAGTTTCGTGAGCGAAACGACAATTTACGTGGGGGAAATCCAATCTTTGAACGGGAAGACAGTTTTCGT---------ACTCGACA---------TTGTGAACGAAATGATAACTTTCGTGAAGCGGAACCATTTAATGATAGC---CCAAGATTTTTGAAAAAATTTCGTAGCCCAAATTTGGACGATTTTGGTCCAAATCAATGCA |
| droGri2 | scaffold_15126:3871458-3871723 + | AAACTTTGATCAGCGAAACGAAAATGTTCGCGATCGAAATGATCC------TTTTCGAAATGACAATTTTCAACTTCGAAACGAAAATTGTCGTGGGCGGCATGAAAACTTTGATAGAGATGACAATATTCATGGGCACGTTGCACGATATGATAATGTTCGTGATCGAAACGATGATTATCGGGAAGTGGACAGATTCAATTATAACCATCCAAGATTTATGGATAATAATCGTAATCCAAGCATGGAAGATATTAATCTTAATCGTCGTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/21/2015 at 04:38 PM