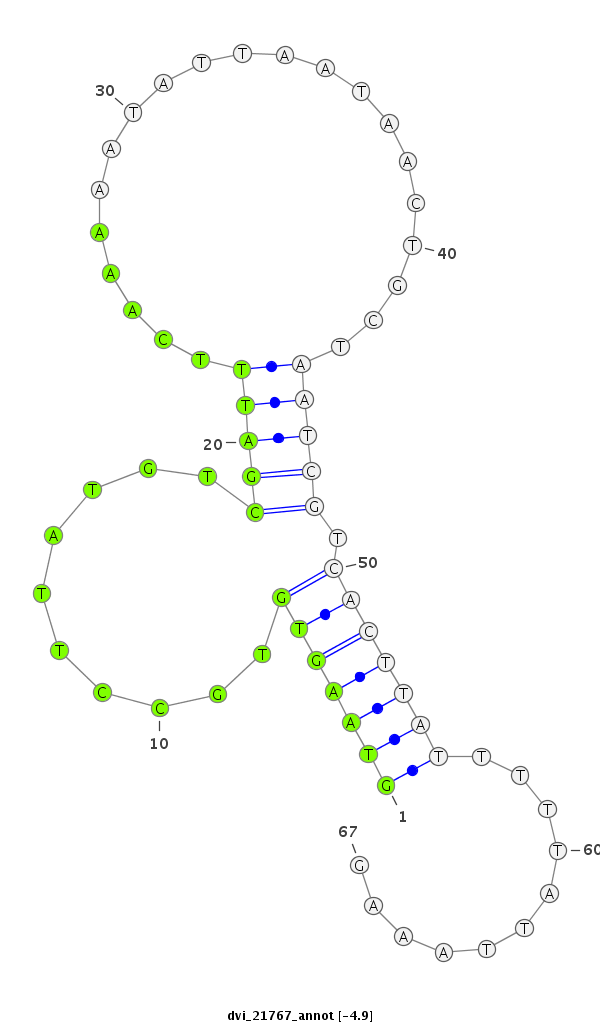
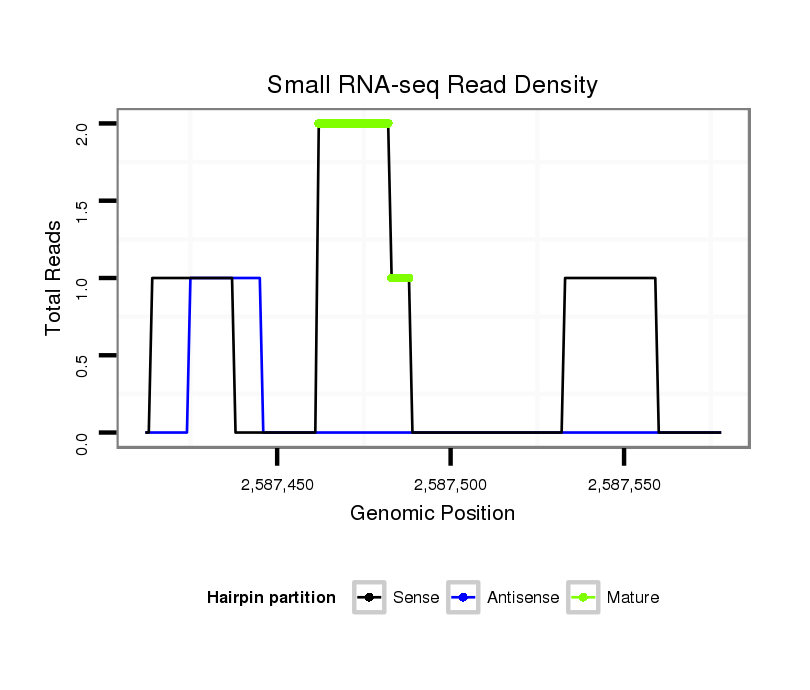
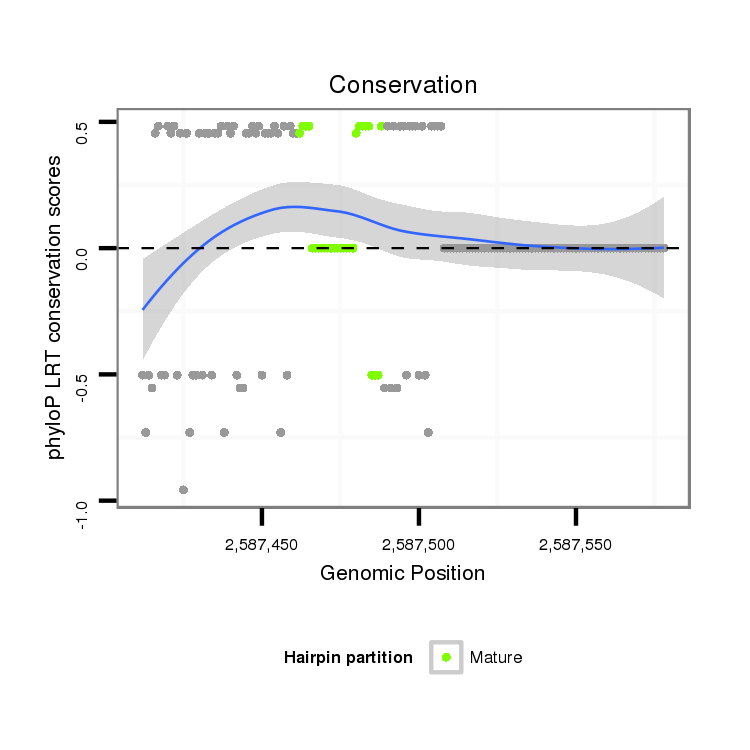
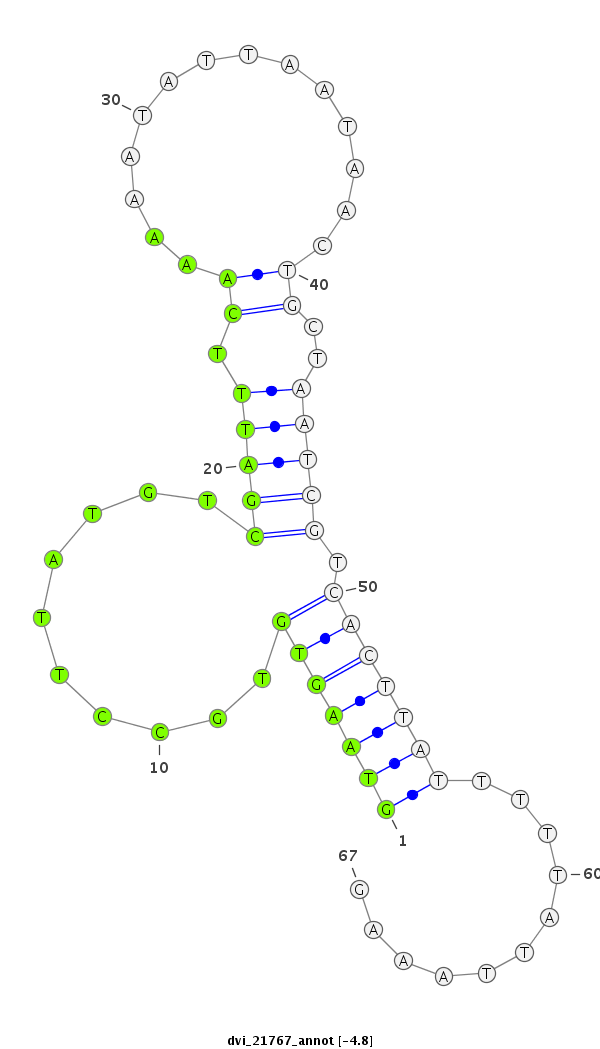
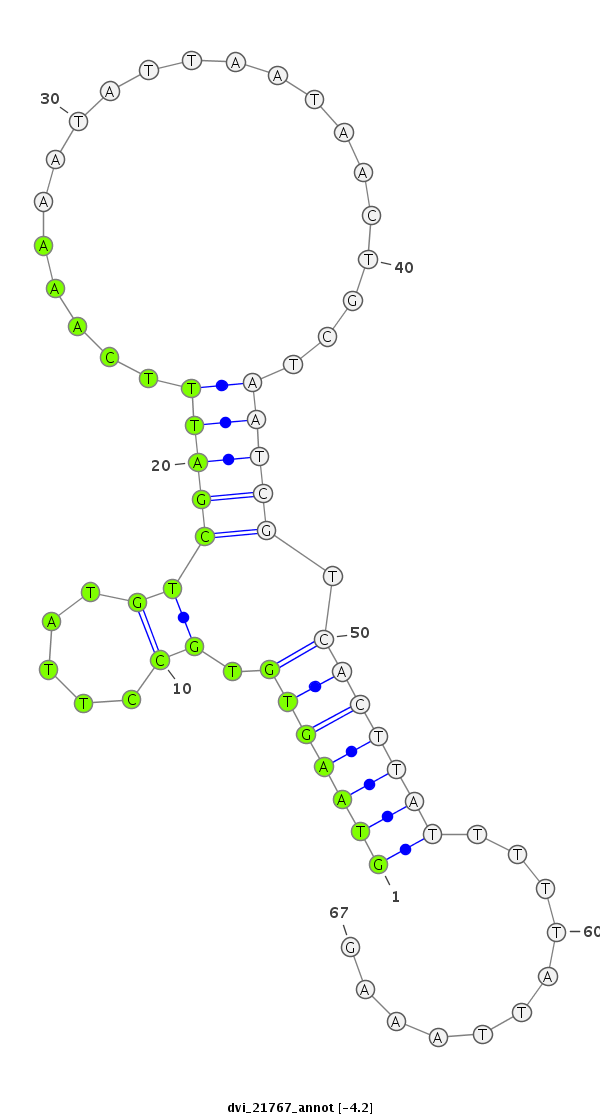
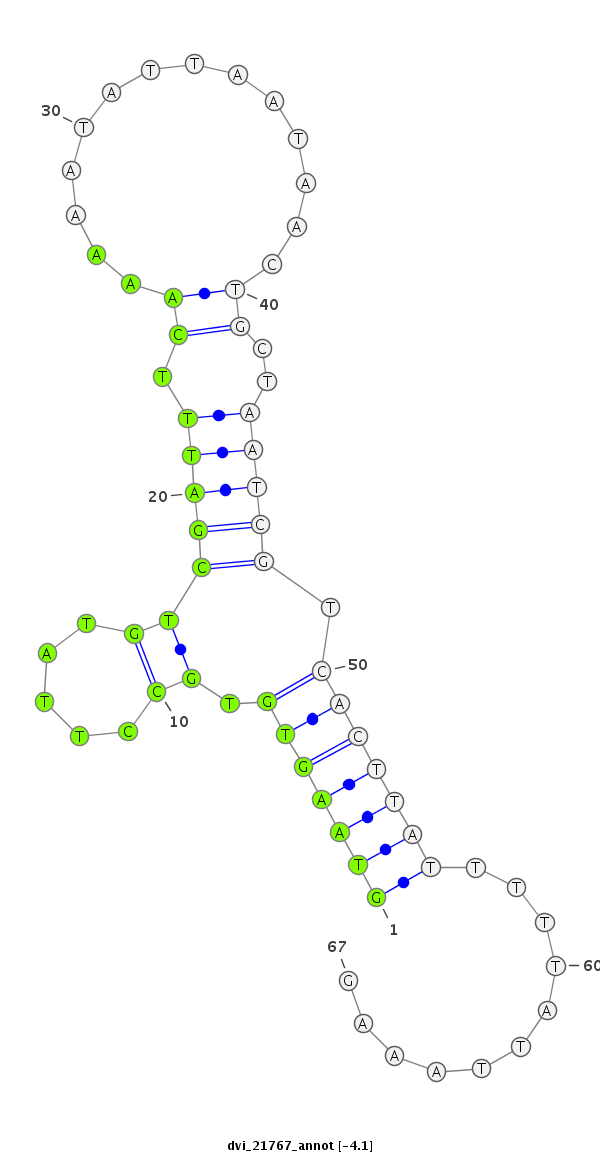
ID:dvi_21767 |
Coordinate:scaffold_13324:2587462-2587528 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -4.8 | -4.2 | -4.1 |
 |
 |
 |
CDS [Dvir\GJ19758-cds]; exon [dvir_GLEANR_5012:1]; CDS [Dvir\GJ19758-cds]; exon [dvir_GLEANR_5012:2]; intron [Dvir\GJ19758-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------################################################## ACGACAATTGTTGGCGCCGTCGCCGACACTGAAGGAGTCGGCAGTAGAGGGTAAGTGTGCCTTATGTCGATTTCAAAAATATTAATAACTGCTAATCGTCACTTATTTTTATTAAAGACGATATGAGAAAGTTGACGAAGACTCCGGCACGCACTGATAAGCATCAA **************************************************(((((((..........(((((.....................))))).)))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
V053 head |
SRR060661 160x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060671 9x160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
V116 male body |
SRR060684 140x9_0-2h_embryos_total |
SRR1106728 larvae |
M047 female body |
SRR060682 9x140_0-2h_embryos_total |
V047 embryo |
M027 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060658 140_ovaries_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................TATGAGAAAGTTGACGAAGACTCCGGC................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GACTCCGGCACGCTCTGAT......... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TGAGAGGGTTGACGAAGACT........................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GACAATTGTTGGCGCCGTCGCCGA............................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGTGTGCCTTATGTCGATTTCAAA.......................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TGAGGGAGTCGGCTGTATAGG..................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CACTGAAGGAGTCGGCAGTAGAGGA.................................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGTGTGCCTTATGTCGAT................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CTACACTGACGGAGTCAGC............................................................................................................................. | 19 | 3 | 11 | 0.82 | 9 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| .............................TGAGGCAGTCGGCTGTAGAGG..................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CTACACTGAAGGAGCCAGC............................................................................................................................. | 19 | 3 | 15 | 0.27 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| .......................CTACACTGACGGAGCCGGC............................................................................................................................. | 19 | 3 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................CGCCTACACTGACGGAG.................................................................................................................................. | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................TACACTGACGGAGTCAGC............................................................................................................................. | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................AAAAACATTATTAACTGCT.......................................................................... | 19 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TATATTAAAGGCGATATAAG........................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GGAGTAGGCATTGGAGGGT................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TGCTGTTAACAACCGCGGCAGCGGCTGTGACTTCCTCAGCCGTCATCTCCCATTCACACGGAATACAGCTAAAGTTTTTATAATTATTGACGATTAGCAGTGAATAAAAATAATTTCTGCTATACTCTTTCAACTGCTTCTGAGGCCGTGCGTGACTATTCGTAGTT
**************************************************(((((((..........(((((.....................))))).)))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060689 160x9_testes_total |
SRR060656 9x160_ovaries_total |
SRR060670 9_testes_total |
M027 male body |
SRR060674 9x140_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................TTCCTCGGCCGTCATATGCC.................................................................................................................... | 20 | 3 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............CGCGGCAGCGGCTGTGACTTC..................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................CGGAACACAGCTAATGTTTT......................................................................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................CTTTCTGCTATACTCTATCA................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................TAAAGTTCTTAAAATTATTG.............................................................................. | 20 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................AAAAAAAATTTTTGCTATAC.......................................... | 20 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........ACCACGGCTGCGGCTCTGA......................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13324:2587412-2587578 + | dvi_21767 | ACGACAATTGTTGGCGCCGTCGCCGACACTGAAGGAGTCG---------------GCAGTAGAGGGTAAGTGTGCCTTATGTCGATTTCAAAAATATTAATAACTGCTAATCGTCACTTATTTTTATTAAAGACGATATGAGAAAGTTGACGAAGACTCCGGCACGCACTGATAAGCATCAA |
| droMoj3 | scaffold_6496:26280534-26280630 - | GAATCAGCTGTCGCCTTTGCCGTCGAAACTATTGGAGTTGCAGAGCAGTCTGGACGCAGGAAAGGGTAA--------------GATTTTGGATAAAATAGTAATTAATAAT----------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 10:03 PM