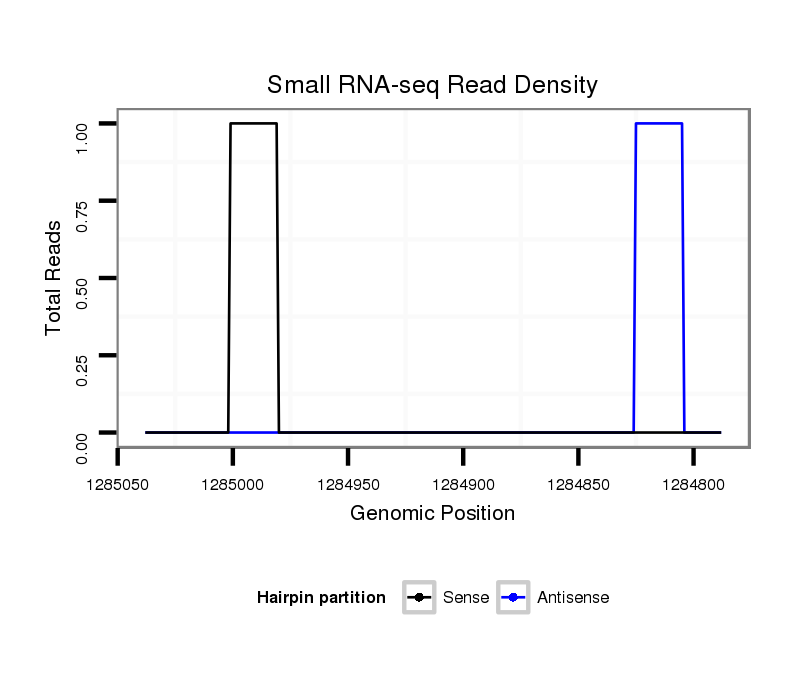
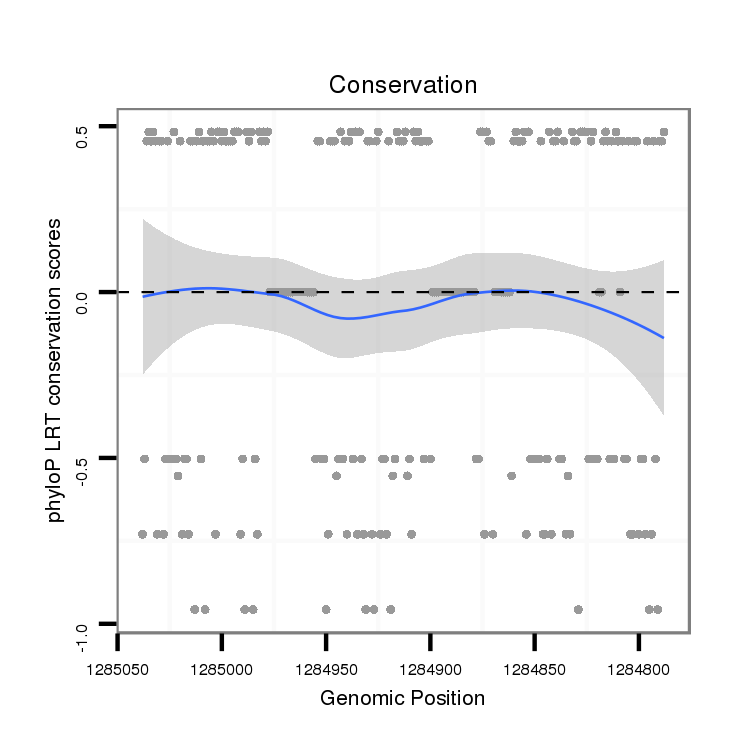
ID:dvi_21305 |
Coordinate:scaffold_13050:1284838-1284988 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18438-cds]; exon [dvir_GLEANR_3114:2]; intron [Dvir\GJ18438-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CGCACTCGGCCAGAGTATGTGCTCGGCTAGCCGAGCTAGAGGCCAAATACACACGGACACTCGAGCAGAGAGCTTAGCTCGCACCCGACCCCGTTAACTGAGATTGGCGGTCGAAATCCGAATCGGATTATGACCCCCACCTGAGCACTTCTTGTCTGCCCGAAGTGCCGGTAATACTGAGCCATAAAATTCATGTTGCAGGCGATAGGGTAAAGGAATCACTGACATCTCGTGTTGCTGACGGTGTCCCA **************************************************...(((.((((((.(((((((((((((((((....((....)).....))))..(((.(((((.((((((...)))))).))))).))).))))))...))...))))).)).)))))))((((((.((((........))))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
SRR1106728 larvae |
SRR060683 160_testes_total |
|---|---|---|---|---|---|---|---|---|
| .....................................AGAGGCCAAATACACACGGAC................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................................................................TTGGCGGTCGATATC..................................................................................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| .........................................GCCAAATACACAAGGACGG............................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
|
GCGTGAGCCGGTCTCATACACGAGCCGATCGGCTCGATCTCCGGTTTATGTGTGCCTGTGAGCTCGTCTCTCGAATCGAGCGTGGGCTGGGGCAATTGACTCTAACCGCCAGCTTTAGGCTTAGCCTAATACTGGGGGTGGACTCGTGAAGAACAGACGGGCTTCACGGCCATTATGACTCGGTATTTTAAGTACAACGTCCGCTATCCCATTTCCTTAGTGACTGTAGAGCACAACGACTGCCACAGGGT
**************************************************...(((.((((((.(((((((((((((((((....((....)).....))))..(((.(((((.((((((...)))))).))))).))).))))))...))...))))).)).)))))))((((((.((((........))))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M028 head |
SRR060687 9_0-2h_embryos_total |
M047 female body |
SRR060679 140x9_testes_total |
SRR1106716 embryo_4-6h |
SRR060672 9x160_females_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060668 160x9_males_carcasses_total |
SRR060666 160_males_carcasses_total |
GSM1528803 follicle cells |
SRR060660 Argentina_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060657 140_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060671 9x160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106718 embryo_6-8h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .GGTGAGCCGGTCTCATGG........................................................................................................................................................................................................................................ | 18 | 3 | 20 | 55.05 | 1101 | 933 | 49 | 32 | 23 | 22 | 0 | 7 | 12 | 0 | 0 | 0 | 5 | 1 | 4 | 3 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| .....................................................................................................................................................................................................................TCCTTAGTGACTGTAGAGCAC................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................CGTTGGGCCAATTGACTCTA................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CGTGAGCCGGTCTCATGG........................................................................................................................................................................................................................................ | 18 | 2 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TGGTGAGCCGGTCTCATGC........................................................................................................................................................................................................................................ | 19 | 3 | 10 | 0.30 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GTGAGCCGGTCTCATGG........................................................................................................................................................................................................................................ | 17 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGTGAGCCGGTCTCATAG........................................................................................................................................................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CCAGTGAGCTGATCTCTCGA................................................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CCAGACGGGCTGCACCGCCA............................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................CGTGTGCTGGGGCAATCTA........................................................................................................................................................ | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................TGACTCTAACGTCCAGGTTT....................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................CCATTACGAATCCGTATTTT.............................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GGGCTAGGGCTATGGACTC..................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGGTGAGCCGGTCTCATGG........................................................................................................................................................................................................................................ | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................TGACTCTAACGTCCAGGTT........................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ACGAGAAGATCGGGTCGATCT................................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................CTGCTGGGGGTGGACTGG......................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13050:1284788-1285038 - | dvi_21305 | CGCACTCGGCCAGAGTATGTGCTCGGCTAGCCGAGCTAGAGGCC---------AAATACACACGGACACTCGAGCAGAGAGCTTAGCTCGCACCCGACCCCGTTAACTGAGATTGGCGGTCGAAATCCGA-------ATCGGATTATGACCCCCACCTGAGCACTTCTTGTCTGCCCGAAGTGCCGGTAATACTGAGCCATAA--AATTCATGTTGCAGGCGATAGGGTAAAGGAATCACTGACATCTCGTGTTGCTGACGGTGTCCCA |
| droMoj3 | scaffold_6680:24661183-24661397 + | AACACTCTGCAGGGATGAGGATGCGCCTGGGCGAGATAGAGGCCCGTTTCCCTAAAGGGACAGATACACT----------------------TCCAGGACCGACAGCGGAAAGTATGGGGGGACGCACCTATCAGAAGTCGGAACCTGACCTCCG---------------------TAAATTGCA--------AGAGCCAGAGTCGGCCCCGATGGCAAACTTGAGGCTAAAAGAGC--CTGGCGTC-CACGGGGCGAGAGCGGCGCCA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/17/2015 at 03:32 AM