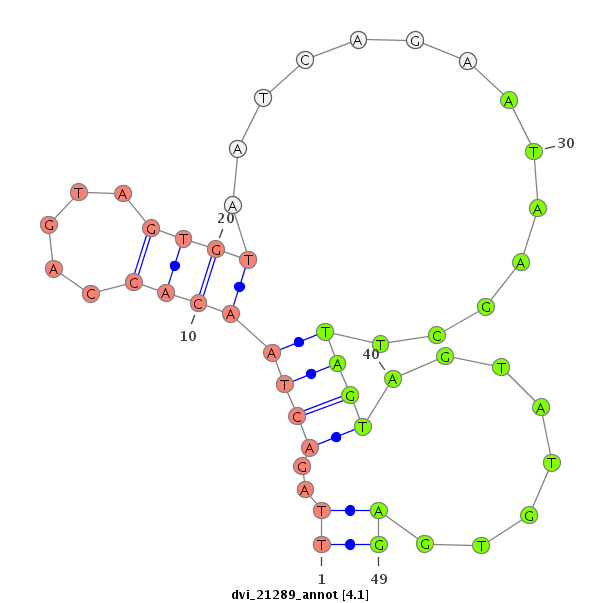
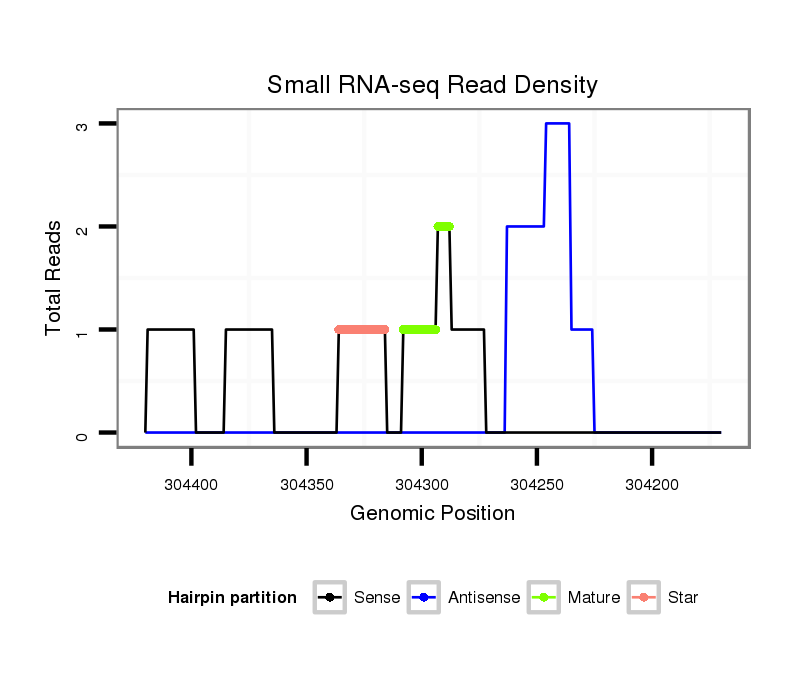
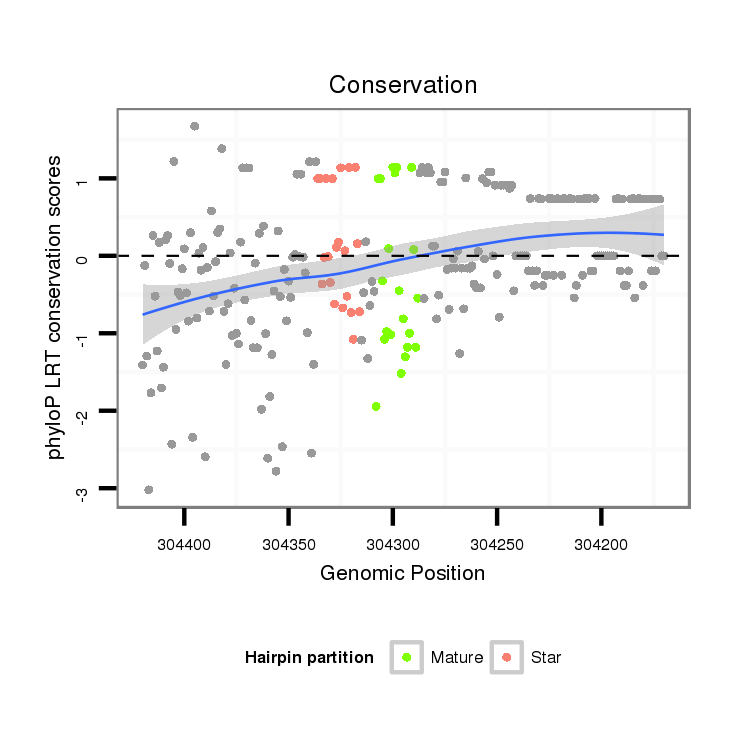
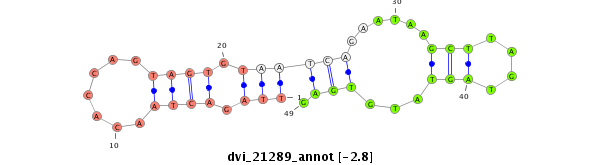
ID:dvi_21289 |
Coordinate:scaffold_13050:304220-304370 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -3.1 | -3.1 | -2.8 | -2.8 |
 |
 |
 |
 |
CDS [Dvir\GJ18451-cds]; exon [dvir_GLEANR_3158:3]; intron [Dvir\GJ18451-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGATGACTGTTGACGAATGGGAGAAGCCAGGACCATCGGAGAGTGACACCCGTCGCGCAACATGTGCACTATACTAAATTTAGATTAGACTAACACCAGTAGTGTAATCAGAATAAGCTTAGTAGTATGTGAGCTGCACTATGGGGATTGAGTATTAACAATTTAGTAATGGATAAGTTAACGAGCAAGCTGAATTTACAGTTGGACCCGGAAATTATTTCAATCCCGTCGGAAGAGGACGCCGGCAGCGT ************************************************************************************((..((((((((.....))))..............))))........))********************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060673 9_ovaries_total |
SRR060675 140x9_ovaries_total |
M047 female body |
V047 embryo |
M027 male body |
V053 head |
SRR1106721 embryo_10-12h |
SRR060684 140x9_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060662 9x160_0-2h_embryos_total |
SRR060671 9x160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................ATAAGCTTAGTAGTATGTGAG...................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GATGACTGTTGACGAATGGGA..................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGTGAGCTGCACTATGGGGAT....................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................TTAGACTAACACCAGTAGTGT.................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TCGGAGAGTGACACCCGTCGC................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTCAAGTCCATCGGAGAG................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.35 | 7 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
| .......................................................................................................................................GCAGTATGGGGATGCAGTAT................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................GTAATCTGAATAAGC..................................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GGCACTATGGGGATTGGCT.................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TGGAAGAGGCCAGGACC......................................................................................................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GGAGTATGGGGATCGAGTA................................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GAACTATGGGGATAGAGGA................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TCGGGGACTGACACCCTT...................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCTACTGACAACTGCTTACCCTCTTCGGTCCTGGTAGCCTCTCACTGTGGGCAGCGCGTTGTACACGTGATATGATTTAAATCTAATCTGATTGTGGTCATCACATTAGTCTTATTCGAATCATCATACACTCGACGTGATACCCCTAACTCATAATTGTTAAATCATTACCTATTCAATTGCTCGTTCGACTTAAATGTCAACCTGGGCCTTTAATAAAGTTAGGGCAGCCTTCTCCTGCGGCCGTCGCA
**********************************************************************************************************************((..((((((((.....))))..............))))........))************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060682 9x140_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060675 140x9_ovaries_total |
SRR060664 9_males_carcasses_total |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TGACCCTCTTCGTTCCTGGTAA...................................................................................................................................................................................................................... | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................TGTTAAATCATTACCTATTCAATTGCTC.................................................................. | 28 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TTCAATTGCTCGTTCGACTTA........................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CACGACGTGATACCCTTAA...................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TTGTACTCGTGATATGAAT.............................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................TATTCAATTGAGCGTTCCACT.......................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................ACACCCGAAGTGATACCGC......................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................TGATACCCTTAGCTCAAAA............................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TTAATGGAGTCATCATACA......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13050:304170-304420 - | dvi_21289 | AGATGACTGTTGACG---AATGGGAGAAGCCAGGACCATCGGAGAGTGACACCCGTCGCGC-----AACATGTGCAC-------------------TA-TACTAAAT-----TTAGATTAGACTAACACCA---GTAGTGTAATCA--GAATAAG-CTTAGTAGTATGTGAGCTGCACTAT-GGG------GATTGAGTATTAACAATTTAGTAATGGATAAGTTAACGAGCAAGCTGAATTTACAGTTGGACCCGGAAATTATTTCAATCCCGTCGGAAGAGGACGCCGGCAGCGT |
| droMoj3 | scaffold_6093:2095-2149 - | ACGGCGACGAGAGCG-----------AG-------------------------------GAAGAGCGGGACGAA-GGCCGAGGGACAATCCCTGAG------------------------------------------------------------------------------------------------CA------------------------------------------------------------------------------------------------------- | |
| dp5 | Unknown_singleton_887:1604-1617 + | CGACCATCGACGAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_4454:1616-1644 + | CGACCATCGACGAAG---AT---G--ACGAAAGGACC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302624:6139-6188 + | AGATGAC-GCCGACAC------CG--AAGCCGGAACCC----------------------ACCAGGAAGACACAGAC-------------------GC----------------------------------------------------------------------------------------------CG------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453385:13938-14077 - | CGGACACCCCCGATAC---GTTTA--ATGACAGCATCGCCGGGCCGATCGACCCGGCGCGGTAAGCAACCAGAA-CC-------------------ATGAAATTAAGTTAAGATCCA---------CACTT---GTAATAGGTTAA--GGT--TAAAAGAGTCCTTTGTTAGCTACACCGT-------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000488056:11255-11384 + | AAGCCACTCCAGACAG---GATCG--ATGACAGCGTCGCCGGACTGAGTAACCCGACACAGTAAGCAGCATGCAGCC-------------------GC-AGATAAATTTAATATTCATTGTATAACCAC--CGTA-----TAACCATTGTATA----------------------------------------------------AGCCTCGTA----------------------------------------------------------------------------------- | |
| droRho1 | scf7180000765460:558-735 + | AGATGACGGACTACC-----------CA------------GGGA--GGCGACACGAGAGGTTAAGCAGGGTAGAAGGTCACCCGGTAATTTCTAAG----------------TTGAATTGTACTAACAC--CCTATGTTATAATCT--AAGTACT-CTAAGTAGAGGTTTCACTGCACCGATTGGTAGGGAGCTTAAACAATAAGGGTCTCGTA-TCTATAAGTT------------------------------------------------------------------------ | |
| droBia1 | scf7180000299209:4763-4949 + | AAATGCA-GCTGCAAAAG-AA--T--ATGACAAGACTACTGGGA--GGCGACACGAGAGGTTAAGCAGGGTAGAGGCTCACACGGTAATTTCTAAG----------------TTGAATTATACTAATACATACAATGTTATTATGT--AAGTATT-CTAAGTCGAGGTTTCACTGCACCGATAGGTAGGGAGCTTGAATAATAAGGGTCTCGTA----------------------------------------------------------------------------------- | |
| droTak1 | scf7180000413188:1161-1394 - | CGGACACCGCCGACCC---GATCG--AGGACAGCACCACCGGGCCAAATAGCCCGGCACGGTAAGCATCTGGAA-CC-------------------ACGGAATAACTTTAAGATCCA---------TGCTT---GTAATAGGTTAA--GAT--TA-ATGAGTCCTTTGTTAGCTACACCGTTAGGTAGGAAGGTCGGAAGGTGGGGTTA---TA-TCAATAAGTC------TAGTTTGCTATTTCAGATGGACGAGGTAATCGT---------GTTGTCGGACGACTCCGACGGC-- | |
| droEug1 | scf7180000409063:12793-12831 + | GGATGAC-GACGACG---ACCACGACATGCCGGGGACACCAGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | v2_chrUn_645:35556-35603 - | AGGAGAATGGTGACG---AGAACGTGGAGTCGGATCCATCGGA-----------------------------------------------------------------------------------------------------------------------------------------------AGGAAGCA------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4929:22017929-22017955 - | TGACGAC-GAAGACC-----------ATGAGAAGAGC-----------------------------------------------------------------------------------------------------------------------------------------------------------CA------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 11:34 PM