
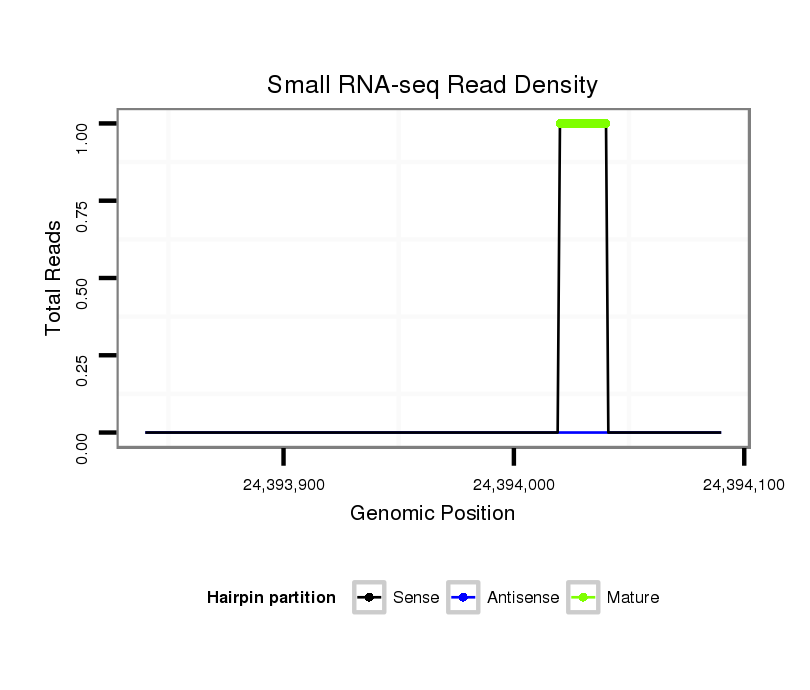
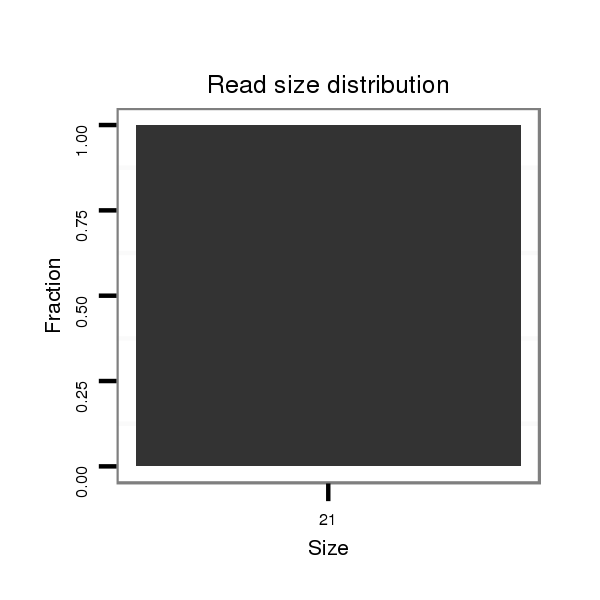
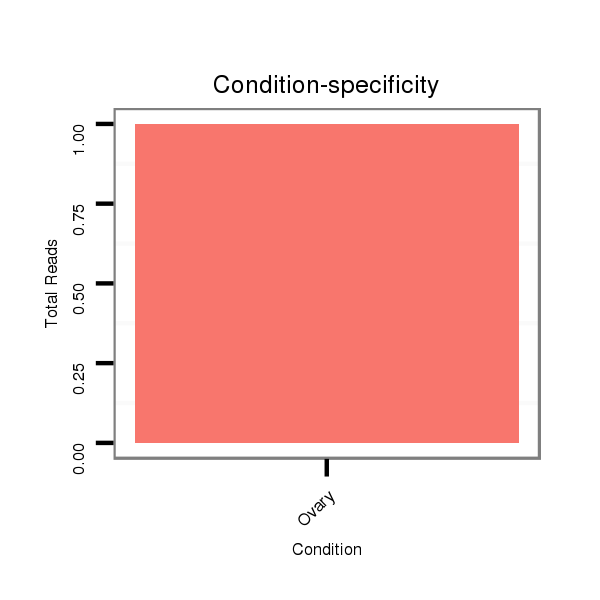
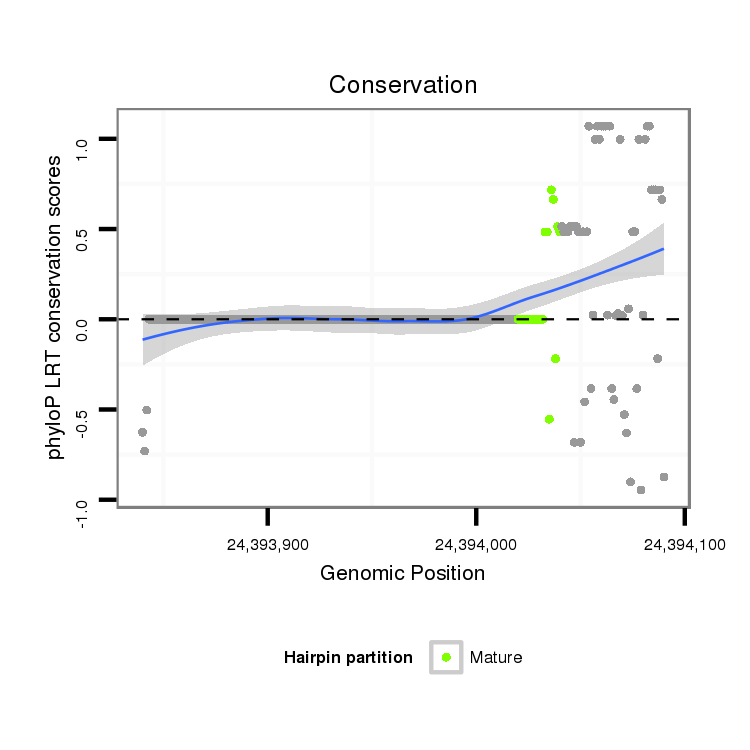
ID:dvi_21254 |
Coordinate:scaffold_13049:24393890-24394040 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.4 | -22.3 | -22.2 |
 |
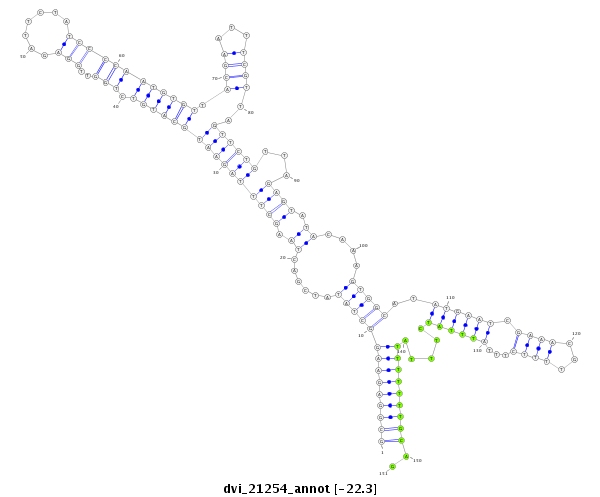 |
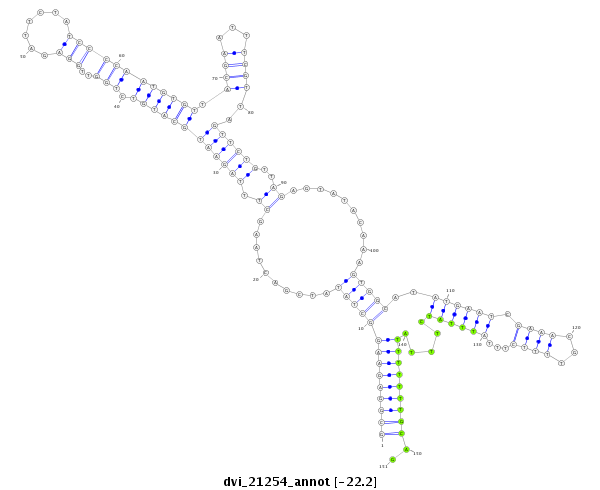 |
exon [dvir_GLEANR_13943:4]; CDS [Dvir\GJ14104-cds]; intron [Dvir\GJ14104-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AACAGTGTTTTTCGCAAATTTATTTGTTAAAAAAGGTCTTTGATAAAGAGGCGGAGAAGGCTATATCGACTAAGCTTTAGAATGCATGTCTGGTTGGAGATTCTATCCCCAATGTGTTACGAATTTCGTTAGTTCTGTTAGAGTATACAAAGTGGCATATGAATCGAAACGTTTTCTTTATTTATCTTTATTTTTTTGCAGACGCATGTGTCCCAAACACAAAATAATACATCGCCCACGACAAATTCAGG **************************************************((((((((((((((......((..((.((((((((((((.(((..(((.......)))))))))))).((((...))))..))))))..))..))......)))))..((((((.((((...))))...)))))).....)))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
SRR060663 160_0-2h_embryos_total |
M047 female body |
SRR060666 160_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................TTTATCTTTATTTTTTTGCAG.................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AGGGGCGGAGAAGTCGATAT......................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................TTTGCTAAAGGGGAGGAGA.................................................................................................................................................................................................. | 19 | 3 | 20 | 0.15 | 3 | 0 | 1 | 1 | 1 | 0 | 0 |
| .............................................................GGTATCGACGAAGCTTTA............................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................TCTTTGGTTATCTTTATGTTTT....................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................GAAGAGGCTGGGAAGGCT............................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................GAAGAGTCGGGGAAGGCT............................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TTGTCACAAAAAGCGTTTAAATAAACAATTTTTTCCAGAAACTATTTCTCCGCCTCTTCCGATATAGCTGATTCGAAATCTTACGTACAGACCAACCTCTAAGATAGGGGTTACACAATGCTTAAAGCAATCAAGACAATCTCATATGTTTCACCGTATACTTAGCTTTGCAAAAGAAATAAATAGAAATAAAAAAACGTCTGCGTACACAGGGTTTGTGTTTTATTATGTAGCGGGTGCTGTTTAAGTCC
**************************************************((((((((((((((......((..((.((((((((((((.(((..(((.......)))))))))))).((((...))))..))))))..))..))......)))))..((((((.((((...))))...)))))).....)))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................CCGTTTTATTATGTAGCGGGT............. | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ..............................................................................................................................................................................................AAAAAAAACTCTGCGTACTCA........................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 |
| ...........................................................................................................................................................................................................................GTATTATTAGGTGGCGGGT............. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:24393840-24394090 + | dvi_21254 | AACAGTGTTTTTCGCAAATTTATTTGTTAAAAAAGGTCTTTGATAAAGAGGCGGAGAAGGCTATATCGACTAAGCTTTAGAATGCATGTCTGGTTGGAGATTCTATCCCCAATGTGTTACGAATTTCGTTAGTTCTGTTAGAGTATACAAAGTGGCATATGAATCGAAACGTTTTCTTTATTTATCTTTATTTTTTTGCAGACGCATGTGTCCCAAACACAAAAT------AATACATCGCCCACGACAAATTCAGG |
| droMoj3 | scaffold_6680:23805355-23805403 - | TCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATGT---------------AGACACAAACTACAAATGTTACATCA---GCAACAAATTTAGT | |
| droGri2 | scaffold_15110:24286935-24286990 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCAGACGCATTTGGCTCAATCACAAAAT------AACACGATGGCCACTTCAAATTCAGG | |
| droWil2 | scf2_1100000004762:2793888-2793922 + | C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGACACAAAATGCTTCTGTTCCAGGG---GCAACAA------C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/16/2015 at 10:52 PM