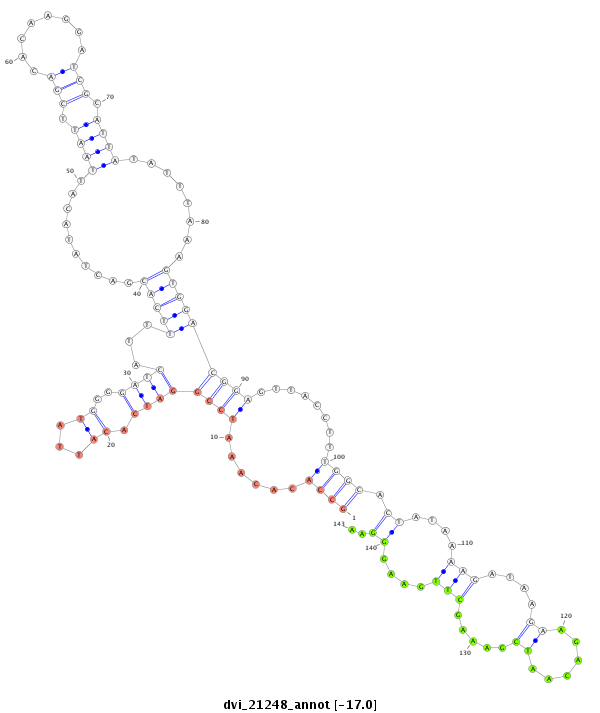
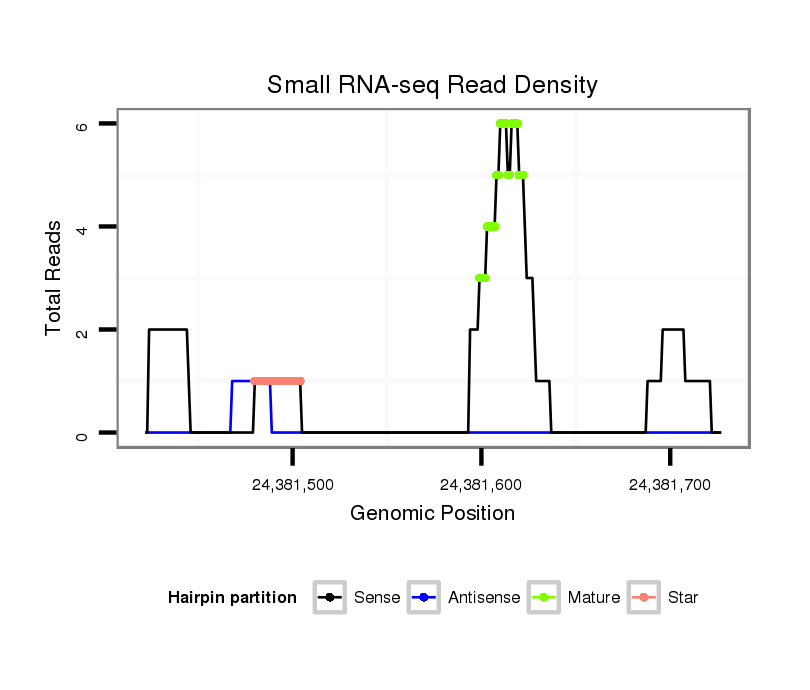
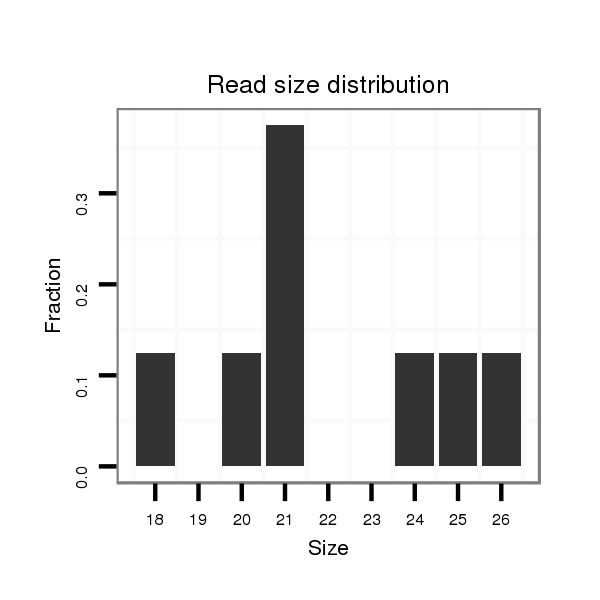
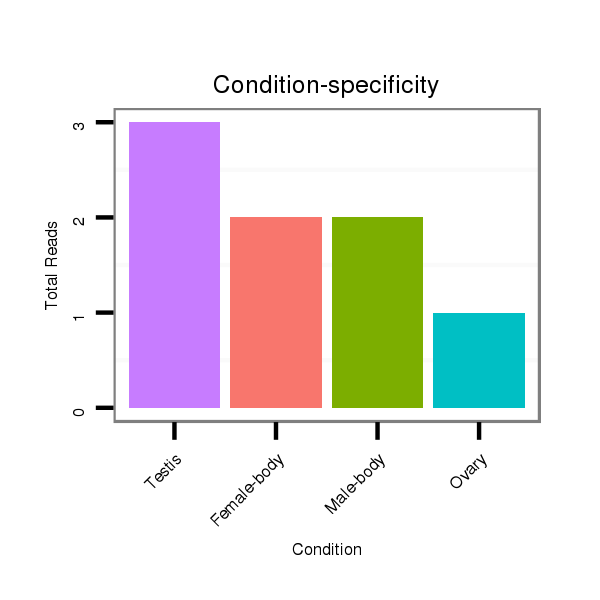
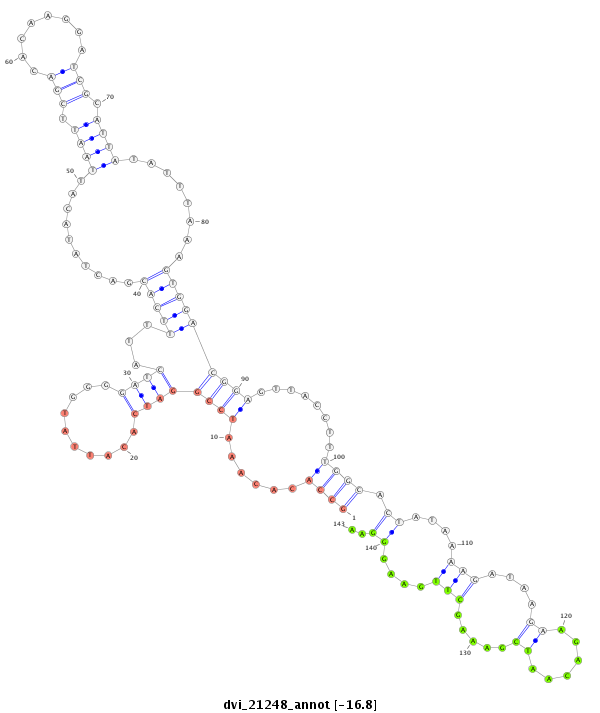
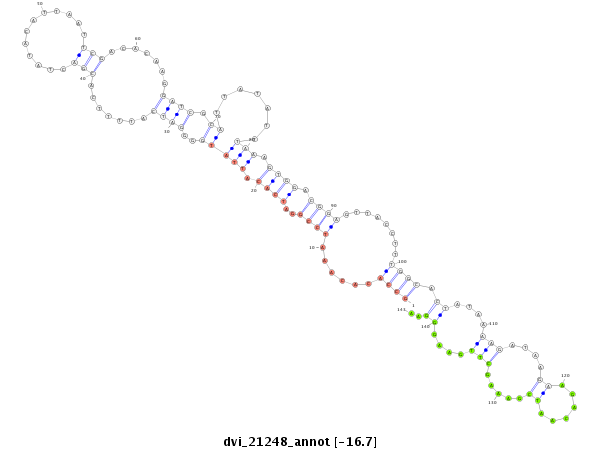
ID:dvi_21248 |
Coordinate:scaffold_13049:24381472-24381677 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
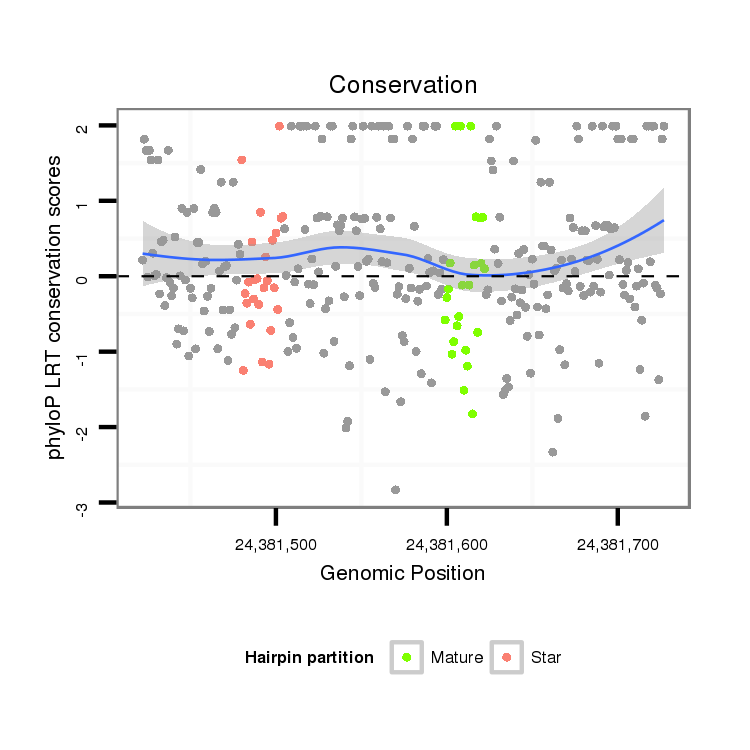
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.8 | -16.7 | -16.7 |
 |
 |
 |
exon [dvir_GLEANR_13941:2]; exon [dvir_GLEANR_13941:3]; CDS [Dvir\GJ14102-cds]; CDS [Dvir\GJ14102-cds]; intron [Dvir\GJ14102-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTTTGGCAGCGTCTTCGGTCAGATAGGTGACGATTATCATGAGGCTACGGTGAATATGCCACACAAATCCGGATCACATTATGGGGATCATTTTCACGACTATACATTAATTCGACACAAGGATCGCATTATATTTAAAGTGGACGGAGTTACCTTTGGCACTATAAAAGATAAGAAGACAATCGAAAGCTTGAAGGGAACTGAGGTAAAAAGCCTTTACGAAAAGTTTACAATTAATAATTAATTATTTTACAGTACTATATCGTATTGGGTTTGACTGCTGGAGGGATATTAAACTTTAAAGA **********************************************************((((......((((((((.((...))..))))...(((((..........((((.(((........))).))))........)))))))))........)))).((....(((....((......)).....)))....))..********************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060678 9x140_testes_total |
SRR060659 Argentina_testes_total |
SRR060670 9_testes_total |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060683 160_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060676 9xArg_ovaries_total |
SRR060687 9_0-2h_embryos_total |
V053 head |
SRR060689 160x9_testes_total |
SRR060656 9x160_ovaries_total |
SRR060655 9x160_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060654 160x9_ovaries_total |
SRR060688 160_ovaries_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................................................TTGGGATTGACTGGTGGA.................... | 18 | 2 | 9 | 3.67 | 33 | 0 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | 4 | 0 | 0 | 0 | 3 | 2 | 3 | 0 | 0 | 1 | 1 | 0 |
| ..............................................................................................................................................................................................................................................................................GCGTTTGACCGATGGAGGGA................ | 20 | 3 | 4 | 1.25 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................AGACAATCGAAAGCTTGAAGGGAA......................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TTTGGCAGCGTCTTCGGTCAGA.......................................................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................AGCTTGAAGGGAACTGAG.................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TATTGGGTTTGACTGCTGGA.................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TTTGGCAGCGTCTTCGGTCAG........................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TAAGAAGACAATCGAAAGCTTGAAGG............................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CTACGTCTTCGGTCAAATAGGT..................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AATCGAAAGCTTGAAGGGAAC........................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AAGGGAACTGAGGTAAAAAGC........................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GCCACACAAATCCGGATCACATTAT............................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AAAGCTTGAAGGGAACTGAGG................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TAAGAAGACAATCGAAAGCT.................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TTGACTGCTGGAGGGATATTAAACTT...... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................GTTTGACCGATGGAGGGA................ | 18 | 2 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................TTGGGATTGACTGGTGGGGGGA................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................TTTGACTGCTGGGGGGAGATTG........... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................TTGGGATTGACTGGTGGAAG.................. | 20 | 3 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................TATGGTATTGGGTTTGAT........................... | 18 | 2 | 17 | 0.24 | 4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................GTTTGACCGATGGAGGGAC............... | 19 | 3 | 17 | 0.24 | 4 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................GTATTGGGTTTAACTGCA....................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................GTTTGAATGATGGAGGGA................ | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................TTGGGATTGACTGGTGGAGA.................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GCAGCGTCATCGGTCAGTCA........................................................................................................................................................................................................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AAAGGGGACGGAGTGACGT...................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TTGAAGGGAACTGCGTTA................................................................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................TGGGTTTGAGTGGTGGAG................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CACCTATCCGGACCACATT................................................................................................................................................................................................................................. | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................................................................TTGGGATTGACTGGTGGAC................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................TACCACATGGTATTGGGTT............................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................CTTGGGATTGACTGGTGGA.................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AAAAACCGTCGCAGAAGCCAGTCTATCCACTGCTAATAGTACTCCGATGCCACTTATACGGTGTGTTTAGGCCTAGTGTAATACCCCTAGTAAAAGTGCTGATATGTAATTAAGCTGTGTTCCTAGCGTAATATAAATTTCACCTGCCTCAATGGAAACCGTGATATTTTCTATTCTTCTGTTAGCTTTCGAACTTCCCTTGACTCCATTTTTCGGAAATGCTTTTCAAATGTTAATTATTAATTAATAAAATGTCATGATATAGCATAACCCAAACTGACGACCTCCCTATAATTTGAAATTTCT
*********************************************************************************************************((((......((((((((.((...))..))))...(((((..........((((.(((........))).))))........)))))))))........)))).((....(((....((......)).....)))....))..********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
M047 female body |
SRR060678 9x140_testes_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................ATGCCACTTATACGGTGTGTT............................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTTTTCGGTAATGCTATTCAG............................................................................. | 21 | 3 | 6 | 0.33 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTTTTCGGAATTGCATTTC............................................................................... | 19 | 2 | 8 | 0.25 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TCCCATGCCACATATGCGG..................................................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................TTTTCGGTAATGCTATTCAG............................................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................ACAAACGACCTCCCTATTAT........... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTTTCGGAATTGCATTTC............................................................................... | 18 | 2 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................AAAAAAATGTCCTGATAT........................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................CCCATGCCAGATATACGG..................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:24381422-24381727 + | dvi_21248 | TTTTTGGCAGCGTCTTCGGTC------------------------AGATAGGTGA-------CGA-TTAT--CATGAGGCTACGGTGAA--------TATGCCACA-------CAAATCCGGATCACATTATGGGGATCATTTTCACGACTATACATTAATTCGACACAAGGATCGCATTATATTTAAAGTGGACGGAGTTACCTTTGGCACTATAAAAGATAAGAAGACAATCGAAAGCTTGAAGGGAACTGAGGTAAAAAG--------C------------------------------C-----TTTA-------------------------CGA-AAAGTTTACAATTAATA---------ATTAATTATTTTACAGTACTATATCGTATTGGGTTTGACTGCTGGAGGGATATTAAACTTT---AAAGA |
| droMoj3 | scaffold_6680:23857824-23858139 - | TTTATGGTAGCGTCTTAAGAAATCAGAC-----------------CG--------AAATGTATGATTTTT---------------------------TAAGGAGCCATAAACTCGAAACGGGGGAACATTATGGACATCATTTTCATAACTATACATTGATACGACATCGGGATCGACTTATATATAAAGTGGATGGGAAAGAATACGGTACAATTGACCATCCCTCGATTTTGGAAGAGATAAATAAAAATGAGATAAATTA--------A------------------------------T-----TTTAAATT----------GCTA-------TATGGTAGGTAAT-------AACCAAG-TCTAGCATTACATTACAGCATTACATCGTACTGGGTTTGACTGCTGGAGGTGTGCTGAATTTCCCCACAGA | |
| droGri2 | scaffold_15110:24266780-24267088 + | TTTTTGGCAGCATTTTCAGTA------------------------ATACAGCCAA-------TGATTTTTTACTCGAGTATTTGTATAC--------CAAGCCACA-------------TGAATCACATTATGGGGAAGCTTTTCACATCTACACTGTGATCAAACATAGAGATCGCATTATTTTCAAAGTAGATGGTGTTACCTATGGCACGATAACGGATAAAATGATTATCGATGATATTAATAAAAATGAGGTATACAT--------T------------------------------T-----ATAGTAACAA----------TGAATTTT----------TTAACAAATAT------G---CTTTTTTTTTTCACAGCACTATATCGTAATTGGTTTGACTGCAGGAGGAACAATAAATTTT---AGGGA | |
| droWil2 | scf2_1100000004762:1492863-1493181 + | TTTTTGGTGGTGTCCTCAATT------------------------GGAATGACAA-------GAA------AGCCGAGCATACACATAC--------TA-GTCGT---------ATGTCCAAACAATCATATGCCAATGGTTTTAATAATTTCACTGTAATTTGGCATCCAGATAGAATTATATATAAAGTAAATGGCCATGACTATGGCCATATAACCGATAAAAAGGCACTGAAGGCGTTTCAAAAACCTG---TAAGTTT--------A------------------------------ACTAAC----------AAATGTTTT-------TTTTTA-AATGAATACAATTAATAACAATGTTTGTTGTTGTTTCTTCAGTGTTTCATTGTTCTGGGCTTGACTGTTCATGGTATTGCCAATTTC---CCAGA | |
| droAna3 | scaffold_13337:18742149-18742472 - | TTTTTGGCAGTGTTTTACATT------GGGGTAAATGTGAATATA----------AATCTGAAGGGTGCGC--------ATTCGATAAATATGTTTCCAAGGAAAG-------------TCAAACAGAATTTTCCAAACATTTTCATAATTACACTATAATTTGGCGCAATGATAAAATCATCTACAAGGTTGATGGCGAAACTTTTGGAATTATTACGGATGAAGTCCTTTTAAAAAAATTCCAAACACAGG---TAACTTA--------A----------------------------------------------AAATATATT-------TTTTTTAATAATTTAA-------AACATA-TTCTTACTCTTTTTTTTAGTGTTATCTAACTCTGGGGTTAACAGTCAATGGTGTGGCAAACTTT---GATGA | |
| droFic1 | scf7180000453824:38848-39114 + | A-------------------------------------------------------------------------------------------------------------------------------CTATGCGGATGGCTTTCACGACTACACAATAATCTGGCAGCCAGATAAAATTGTATTCGAGGTCGATGGAGAATTTTTCGGGGCCATTACAAATGCAACACTCCTTGAACCCTTTAATAAGCATGAGGTGAGTACAATTGTAAGCTTTTTGTAAAAGCATGATTTAATTGCATAT-----TTTTTGGTAT----------TGCTTATT----------TTAATATTCCT--------------GTTTTATTTTAGTGTCGTCTAGTCCTGGGCTTAACTGCTGGTGGAAATGTTAATTTT---AATGA | |
| droEug1 | scf7180000409471:812747-812978 - | AT-----------------------------------------------------------------------------------------------------------------------------ATTACGCTGACGCCTTTCACGACTATTCTATTATCTGGCAGGAGGATAAAATTGCCTTTAAGGTCGATGGGGAGTTTTTCGGAGCCATAACCAATGCAACGCTTCTTAAGGCATTCAACAAACACGAGGTAAAGCC--------A------------------------------T-----GTTT-AAA----------AGTTATTATT-------GTTTC----TTAT-----------TTTTTTCATGTTTTAGTGTCATATAGTCCTGGGCCTAACTGCTGGAGGAAATGTTAATTTC---AATGA | |
| droSim2 | 3l:18306500-18306805 + | TTTATGCCGGGGCAATCCTTAATGATTATGGAACACGGACATACG----------AATTGGAGGAGTTTG---------------------------CCAGTAGC---------------GACGATCACTATGGGAATGACTTTCACACTTACACGGCTATTTGGAAGGAGGAGAGCATTAATTTTAAGATAGATGGTCGCACCTTTGCTCAAATAACCAATTCAACGATCGTTCAAGAACTAAGTAAAACAGAGGTAAGTTT--------A------------------------------C-----TTAATAAT----------ACCAATTGTT-------TTGCA----TTAA-----------AATTCCAACTTCTCAGCGCCATATCGAATTGTTCCTAACTGTCGGCGGAGAGTACAATTTT---CCGGA | |
| droEre2 | scaffold_4784:15243252-15243493 - | T--------------------------------------------------------------------------------------------------------------------------------TATGGGGACGATTTTCACGACTACACTATTATCTGGCAGACTGATAAAATTATCTTTAAGTTTAATGGCGAGTTCTTTGGAGCAATACACAATGCGACTCTTCTTGAACCATTCAATAAACATGAGGTGAGACC--------GCTCTCTC------------------------------TAAAAAT----------G-------TTATTTAAAGTTTTAA-------GAACTC-TAGGATCTTTCGTTTTCAGTGCCATCTAGTGCTGGGCCTAACCGCCGGCGGAAATGTAAATTTT---AATGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 10:41 PM