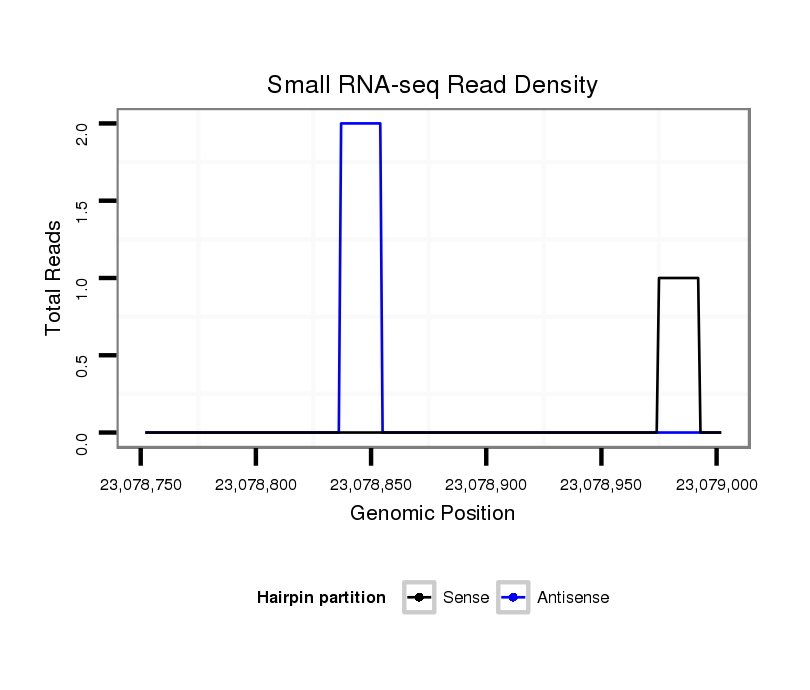
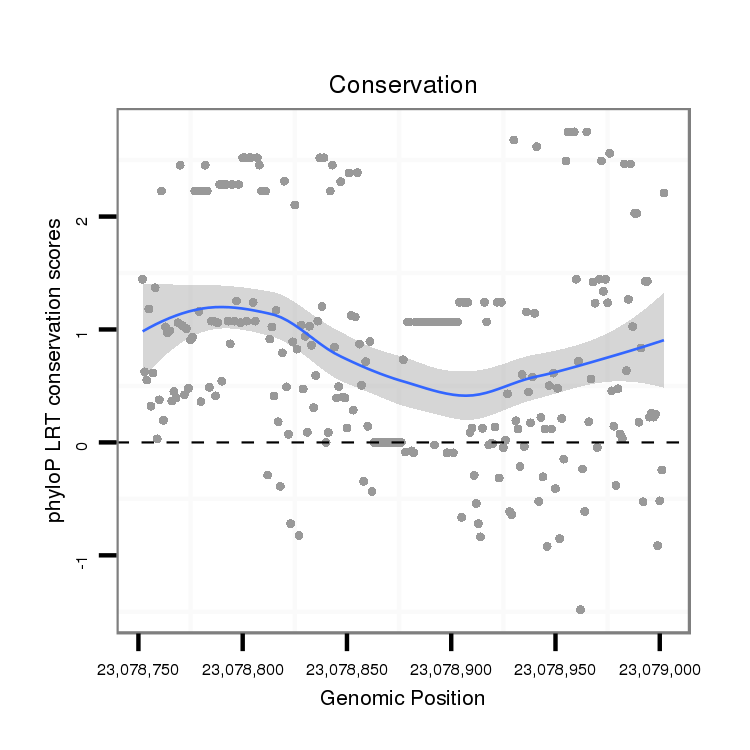
| droVir3 |
scaffold_13049:23078752-23079002 + |
dvi_21081 |
TGGCTGAAGCAGCTCT------GCAGCGGCAGCAACAGGC---GGCGACCGTCGACGATGTAAGTAGCGCCC-GTAAC--TCCT---CCCAATCAC--------AAAGTCAAC-------CACC---AATCCAA-----CA-----ATTATA-TATTGCTATATATACAATTTCCATATATATATATATACATGTATATATATTGCCCTATTTGTTTT-----------TACGTGTATTTGTT-----------------------------T--------TCT-TTGGCAGCT--G----GCAAAATTTT----------GAGC--AC-CTCT-TCAC-CTAAGAATTGGCTA----A-AAAAAATAT-A |
| droMoj3 |
scaffold_6680:17690293-17690514 + |
|
TGGCTGAAGCAGCTCT------GCAGCGGCAGCAACAGAC---GGCGACCGTCGACGATGTAAGTAGCGCCC-AA--G--TAAC--TCCCAATCAC--------AAAGTAA-AC------CA-ATCCT-TAAAATCAACCA-----ATTAGA-TC------------------------------------------------------------------T-----CTCTATTGCCA---CTTTGTTTTTATG-TGCA---TT-TG-GTTTT--------TCT-TAAGCAGCG--G----GCAAAATTAT--G-------TAGC--AC-CTCTCTAAC-TACAAAATTGGCAAAGAAC-AAAAAACGC-A |
| droGri2 |
scaffold_15110:18346306-18346523 + |
|
TGGCTGAAGCGGCTCT------CCAGCGGCAGCAACAGGC---GACGACCGGCGACGATGTAAGTAGCGCCC-GT------ATC-----------C--------AAAGTAA-ATCAAACCCAAAAAGAAAAAAATATATAT-----AAATGTATT-----------------------------------------TTTATGCCAATTT----GTTTTTACG-----TTTATGTA---------------------------TC-TG-TTTATTCTTTATGTAG---TGTGGC---AGCAATCAAAATTA-----------TAGCTCAC-CTCT-TAAC-AGCAGAATTGGCAA----CA---------AA |
| droWil2 |
scf2_1100000004822:55752-55889 - |
|
TATACAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTCTATATATATATATATATATATATGTATACTGAGCTATTTATATA-----------TATATATATACACC-----------------------------T--------TGT-AAAACATAG--T----CCAAAATGTTACCTAGCTAACATA--TC-ATCT-TAAA-CACATAGTTA--------A-AAAAAAAAA-A |
| dp5 |
XR_group8:2405380-2405600 + |
|
TGGCCGAGGCCGAGGCAACTCCGCAGCACGAGCGACAGGC---GACGACCGGCGACGATGTAAGTAGCGAGC-GTT-G--TTAA---CCTCCTATC-------AAAAGTAA-C-------CACCTCCCAGCAAATTTCCAA-----AATCGTTTT-----------------------------------------------------------------CG-----TTTGCGTGCCT-TATTTCGTTATCGTT-TGTT---TTTAT---TGA--------TATTTTGGTAGCAGAG----ACAAAATTG-----------TATT--AC-CTCT-TAAC-AGCAGAATTGGCAA----C----------AA |
| droPer2 |
scaffold_24:649249-649466 - |
|
TGGCCGAGGCCGAGGCAACTCCGCAGCACGAGCGACAGGC---GACGACCGGCGACGATGTAAGTAGCGAG----C-G--TTAAC-CTCCTATCA---------AAAGTAA-C-------CACCTCCCAGCAAATTTCCAA-----AATCGTTTT-----------------------------------------------------------------CG-----TTTGCGTGCCT-TATTTCGTTATCGTT-TGTT---TTTAT---TGA--------TATTTTGGTAGCAGAG----ACAAAATTG-----------TATT--AC-CTCT-TAAC-AGCAGAATTGGCAA----C----------AA |
| droAna3 |
scaffold_13337:11919282-11919475 + |
|
TGGCTGAGGCGACGGT------GCAGAGTGCGCACCAGGC---GACGACCGGCGACGATGTAAGTAGCGGCCAATC-GTGTAATCCACCCAACCAC--------CCAGTAA-CCAA---CCACCTCCAAGCAAT--------------------------------------------------------------------------------------------------ATGCCATTGAC---TTATTATT-TGTT---TTTTA-GT---------------------TCAGCAGC--TCAAAATTTCTACCTAC---CAGC--ACCTTC--TAACCAGCAGAATTGGC------------------- |
| droBip1 |
scf7180000396589:2068603-2068800 - |
|
TGGCTGAGGCGACGGT------GCAGAGTGCGCACCAGGC---GACGACCGGCGACGATGTAAGTAGCGGCCCATC-G--TAAT---CC-CATCCA-------TCCAGTAA-CC-A---CCACCTCCAAGCC--TTTCGAA-----AAT-----------------------------------------------------------------------------------ATGCATTTGACTCGTTATTATT-TGTT---TT-AA-GT-----------TTC-ATTGTAGCTGCA----GCAAAATTT-----------CAGC--ACCTTC--TAACCAGCAGAATTGGCC------------------ |
| droKik1 |
scf7180000302391:830275-830500 + |
|
TGGCTGATCCGGCGGC------GCAGAGTGCGCACCAGGCGGCGACGACCGGCGACGATGTAAGTAGCGTCCCATC-G--TAAC--ACCCAATCAC----------CGTAA-C-------CACCTCCAAGCAAATTATCCA-----AATCGTTTC-----------------------------------------------------------------TGCATATTATATGTGCCTTTGTTTTGTTATTGTT-TGTCTTTTTTTG-GTTAT--------TCT-T---------CA----GCAAAATTTG----------TAGC--AC-TTC--TAAC-AGCGAAATTGGCCA----A-AACTCGATT-A |
| droFic1 |
scf7180000454113:2654451-2654653 + |
|
TGGCTGATCCGGCGAC------GAAGAGTGCGCACCAGGC---GGCGACCGGCGACGATGTAAGTAGCGCCT-ATCCG--TAAT---CCCCATCAC-------TCCAGTAA-C-------CACCTCCAAGCAAATTTCCAA-----AATC----------------------------------------------------------------------------------GTGCCTTTGTTTCGTTTTTGTT-TGAT---TTTTC-GTTGT--------TCT-TTCGTATCTGCA----GCAAAATTG-----------TAGC--AC-TTC--TAAC-AGCGAAATTGGCCG----------------A |
| droEle1 |
scf7180000491277:507860-508034 + |
|
AACCATAAA-----------------------------AT---AAAGCCCAGCATCTATATACAT---------------------------GCAC-------ACATATAA-A--------------------------------------------------------ATTTATATATATATAAATATATATATATATATACCTCTCTAACCGTGTA-----------TATATGTATATG-------ATCGAT-TGTA---TTTTT------------------CAAAGTTCA-AA----GCAAAAAAT-----------AAAC--AA-AAC--AAAA-AAAAA-TG----CA----A-AAAAAATAT-A |
| droRho1 |
scf7180000769020:84437-84627 + |
|
TGGCTGATCCGGCGGC------AAAGAGTGCGCAACAGGC---GACGACCGGCGACGATGTAAGTAGCGCCC-ATC-G--TAAT---CCCCATCAC-------TCCAGTAA-C-------CACCTCCAAGCAAATTTCCAA-----AATCGATTC------------------------------------------------------------------T-----TTCACGTTCCTTTGTTTCGTTATTGTT--------ATTAT-TTTCT--------TCT-TTCGTATCAGCA----GCAAAATTT-----------CAGC--AC-TTC--T----------------------------------- |
| droBia1 |
scf7180000302428:4237350-4237565 - |
|
TGGCTGATCCGGCGGC------GAAGAGTGCGCACCAGGC---GACGTCCGGCGACGATGTAAGTAGCGCCC-ATC-G--TATT---CCCCATCAC-------TCCAGTAA-C-------CACCTCCAAGCAAATTTCCAA-A---AATCGTTTT------------------------------------------------------------------T-----TTCACGTGCCTCTGTTTCGTTATTTTTATGAT---TTCCC-ATTGT--------TCT-TTCGTAGCCGCA----GCAAAATTG-----------TAGC--AC-TTC--TAAC-AGCAAAATTGGCCA---------------AA |
| droTak1 |
scf7180000415254:634014-634214 + |
|
TGGCTGATCCGGCGGC------GAAGAGTGCGCACCAGGC---GACGTCCGGCGACGATGTAAGTAGCGCCC-ATC-G--TAAT---CCCCATCAC-------TCCAGTAA-C-------CACCTCCAAGCAAATTTCCAA-A---AATCGTTTC----------------------------------------------------------------------------------TTTGTTTCGTTACTTTT-TGAT---TTTTC-GTTGT--------TCT-TTCGTATC---A----GCAAAATTG-----------TAGC--AC-TTC--TAAC-AGCGAAATTGGCCA---------------AA |
| droEug1 |
scf7180000409466:4083587-4083786 - |
|
TGGCTGATCCGGCGGC------GAAGAGTGCGCACCAGGC---GACGTCCGGCGACGATGTAAGTAGCGCCC-ATC-G--TAAT---CCCAATCAC-------TCCAGTAA-C-------CACCTCCAAGCAAATATCCAA-AAAAAAAAGTTTC------------------------------------------------------------------T-----TTCACATGCCTTTGTTTTGTTATTGTT-TGAT---TTTTTCGTTCT--------TCT-TTCGTACAAGCA----GCAAAATTA-----------TAGC--AC-CTC--T----------------------------------- |
| droSim2 |
3l:11693924-11694135 - |
|
TGGCTGATCCGGCGGC------GAAAGGTGCGCACCAGGC---GACGTCCGGCGACGATGTAAGTAGCGGCC-ATC-G--TAAA---CCCCATCAC-------TCCAGTAA-C-------CACCTCCAAGCGAATTTCCAAAA---AATTGTTAC------------------------------------------------------------------C-----TTCACTTGCCT-TGTTTCGATATTATT-TGAT---TTTAT-GTTGT--------TTC-CTCGTATCAGCA----GCAAAATTC-----------GAGC--AC-TTC--TAAC-ATCGAAATTGGCC------------------ |
| droYak3 |
3L:12040255-12040473 - |
|
TGGCTGATCCGGCGGC------GAAAGGTGCGCACCAGGC---GACGTCCGGCGACGATGTAAGTAGCGCCC-ATC-G--TAAA---CCCCATCTCACTCCTGTCCAGTAA-C-------CACCTCCAAGCAAATTTTCAA-A---AATCGTTTC------------------------------------------------------------------C-----TTCACTTGCCT-TGTTTCGATATTATT-TGAT---TTTAT-GTTGT--------TCC-CTCGTATCAGCA----GCAAAATTT-----------GAGC--AC-TTC--TAAC-ATCGAAATTGGCCA----------------- |
| droEre2 |
scaffold_4784:11981813-11982025 - |
|
TGGCTGATCCGGCGGC------GAAAGGTGCGCACCAGGC---GACGTCCGGCGACGATGTAAGTAGCGCCC-ATC-G--TAAT---CCCCATCAC-------TCCAGTAA-C-------CACCTCCAAGCAAATTTTCAA-A---AATCGTTTC------------------------------------------------------------------C-----TTCACTTGCCT-TGTTTTGATATTATT-TGTT---TTTAT-GTTGT--------TCC-TTCGTATCAGCA----GCAAAATTT-----------GAGC--AC-TTC--TAAC-ATCGAAATTGGCCA----------------A |