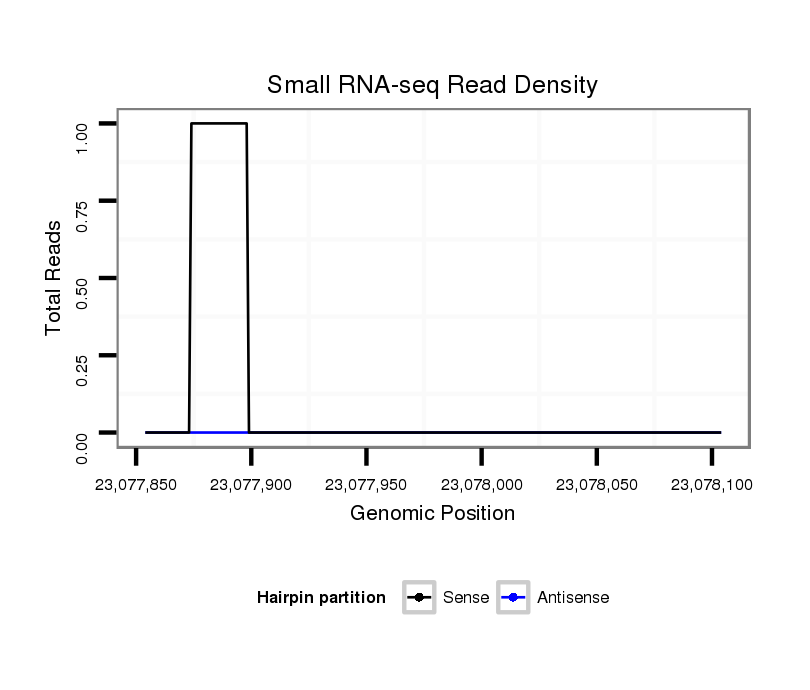
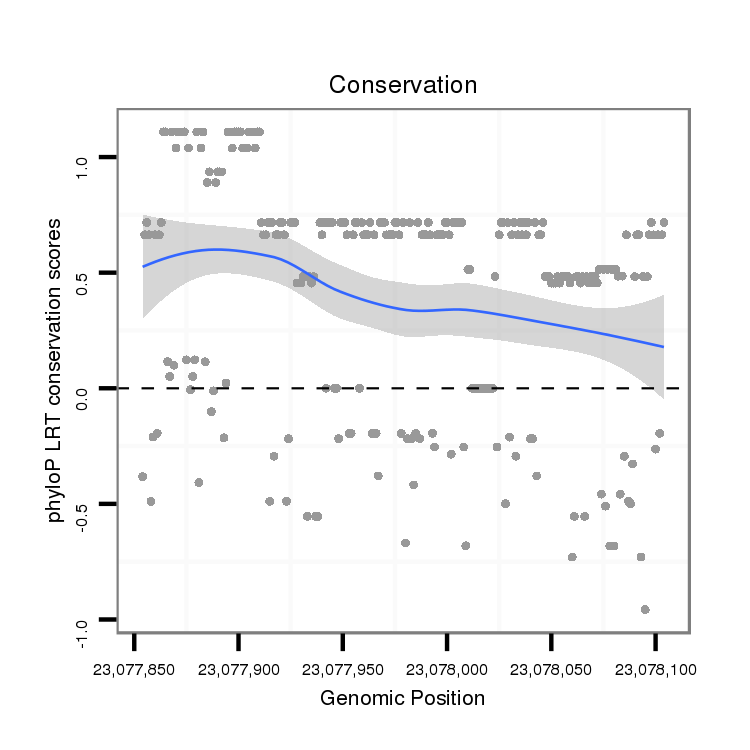
ID:dvi_21079 |
Coordinate:scaffold_13049:23077904-23078054 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_13860:1]; CDS [Dvir\GJ14014-cds]; intron [Dvir\GJ14014-in]
No Repeatable elements found
| -----------------------------------------#########--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACAGCAGCCAAAAAAGCATTTTGTAAAACAACAAGCATACTATGAAATGGGTAAGTTTCCATAGGCAAGGCAATGCCAATTCATATGTATATTTTTTAGGCCATTTGGCAGGCATAAGGGCTTCCTGGTTTCAGCGGTGGTGGCCTTCAATTTTTTTTTTCTTTTTCTTATGTTAAACTGCACACATTTTCCATATGCGTGTATTGCATACATACACACTCTATGTGTGTGGCGAGTGGCTGTCTCACGCA **************************************************...(((((.((((((...(((...)))....................((((((((..((.((.((.(((....))).)).)).))...))))))))....................))))))..))))).(((.(((.......))).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
V116 male body |
M061 embryo |
M047 female body |
SRR060667 160_females_carcasses_total |
SRR060656 9x160_ovaries_total |
SRR060673 9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................TTGTAAAACAACAAGCATACTATGA.............................................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................TAAGAAATGGGAAAGTTTC................................................................................................................................................................................................ | 19 | 2 | 6 | 0.33 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................TGAAGTGGGGAAGGTTCCA.............................................................................................................................................................................................. | 19 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................CTATAAAATTGGTAAGTTCCC............................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................AAGAAATGGGCAAGTTTCCAAA............................................................................................................................................................................................ | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................TGAAGTGGGAAAGGTTCCA.............................................................................................................................................................................................. | 19 | 3 | 16 | 0.19 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TGAATAGGGTTAGTTTCCA.............................................................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ..............................................................................................................................................GCCTTCAATTTTTTATTTTTT........................................................................................ | 21 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CACTCTAAGCGTGGGGCG................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGTCGTCGGTTTTTTCGTAAAACATTTTGTTGTTCGTATGATACTTTACCCATTCAAAGGTATCCGTTCCGTTACGGTTAAGTATACATATAAAAAATCCGGTAAACCGTCCGTATTCCCGAAGGACCAAAGTCGCCACCACCGGAAGTTAAAAAAAAAAGAAAAAGAATACAATTTGACGTGTGTAAAAGGTATACGCACATAACGTATGTATGTGTGAGATACACACACCGCTCACCGACAGAGTGCGT
**************************************************...(((((.((((((...(((...)))....................((((((((..((.((.((.(((....))).)).)).))...))))))))....................))))))..))))).(((.(((.......))).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060676 9xArg_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060671 9x160_males_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................AAAAAAAAAGAAAAGGGATACCAT............................................................................ | 24 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...CGTCGGTTTTTTCCCAGAACA................................................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................ACCGGAAGTTAAAAAGAAGAGAC........................................................................................ | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................CCGGAAGTTAAAAAGAAGAG.......................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................ATGTGCGAGATACACCCA..................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CGGAAGTTAAAAAGAAGAGA......................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................CGGAAGTTAAAAAGAAGAG.......................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................CACCACCGGAGGTTAATAT................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:23077854-23078104 + | dvi_21079 | ACAGCAGCCAAAA-------AAGCATT-TTGT--AAAACAA-CAAGCATACTATGAAATGGGTAAGTTTCCATAGGCAAGGCAATGCCAATTCATATGTATATTTTT------------TAGGC-----CATTTGGCAG-------GCATAAGGGCTTCC-----T---GGTTTCAGCGGTGGTGGCCTTCAATTTTTTTTTTCTTTTTCTTATGTTAAACTGCACACATT------------------------TTCC--ATATGCGTGTATTGCATACATACACAC-----------------------------TCTATGTGTGTGGCGAGTGGCTGTCTC-------------------------ACGCA |
| droMoj3 | scaffold_6680:17689428-17689698 + | GCAGCAGCCAAAA-------AAGCATTTTTGTAAAAA-CAACCAAGCATACTATGAAATGGGTAAGTTTCCAGAGGCAAGGTAATGCCAAATCAATTGT-TAT--TT------------TAGGC-----CAT-TGGCAG-------GCATAAGGGCTTCC-----T---TATTACAACGGTGGCGGCCTTCGATTTTC--------------ACGTTCAACTGCACACATT------------------------TTCCGCATATGCGTGTATTGATTACAAACACACTTGCATTGAATGTGTAGATGTGT------------------GCGCTTGGATCTCTCTCTCTTTCTCTCAAA---CACACGTTCGCA | |
| droGri2 | scaffold_15110:18345439-18345731 + | GCAGGGGTCAAAAGCGCAAAAAGCATT-TTG--AAAA-CAA---------TCATGAAATGGGTAAGTTTCCAGATGCAAGCCAAT-----------TGT-TAT--ATACAAAATGTAAATAGATGTTCACAT-TGGCAATGTTCTTGTCTAAGGGCTTCTCTTCCTGTGTGTAATAGCGGTGATGGCCTTCGATTTTTGTT------------TGTTAAGCTTCACACAAACACACACACACACATCTCTCTCCATGCC--A----------------------------------AATGTGTGCATGTGTATGCGTTTTTTTTTTGCGTCCAG-GG----CTCACGCATGCACTTAAATGACACTCACACACA | |
| droKik1 | scf7180000302391:829313-829361 + | A---------AAT-------CAACATTGTCGC--CGACCAT-CACACATATTATGAAATGGGTAAGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/21/2015 at 04:22 PM