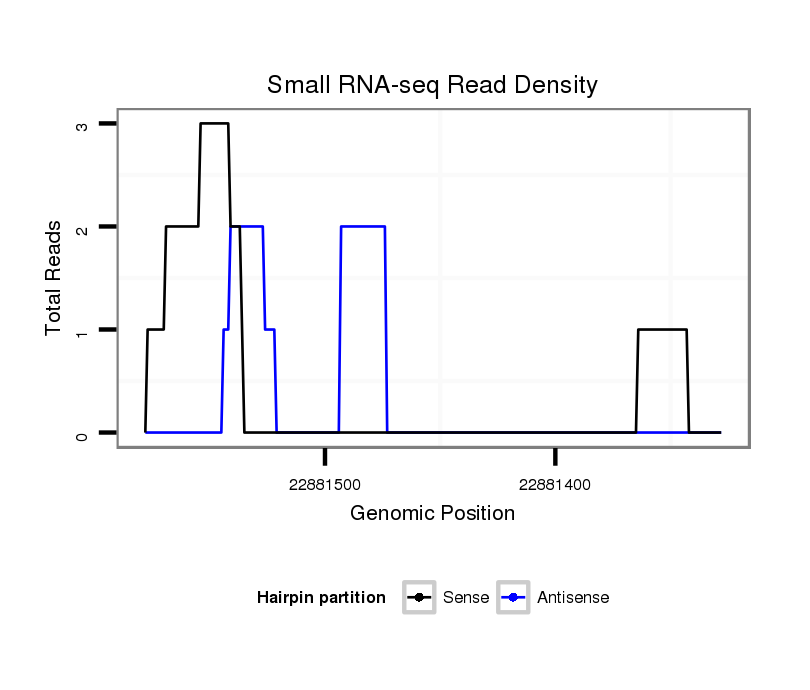
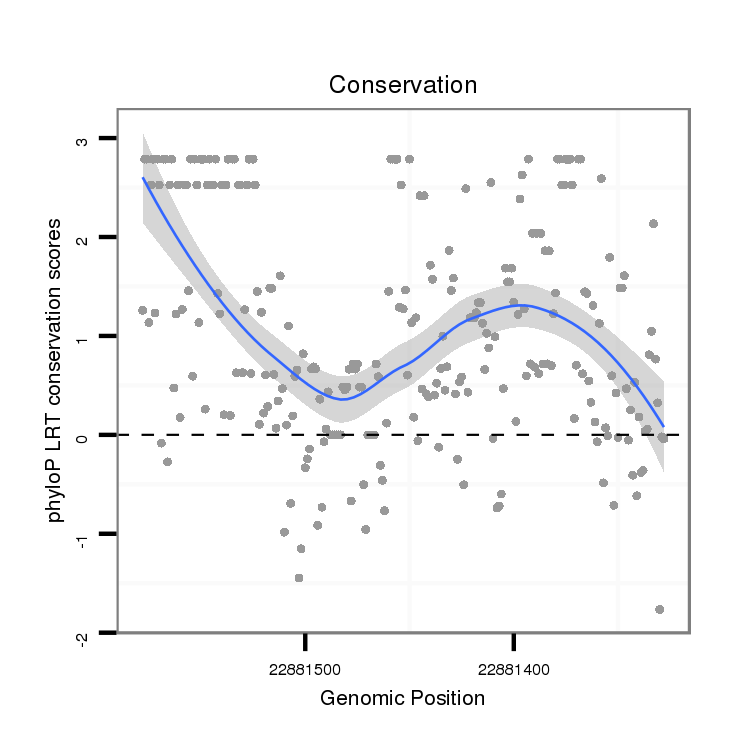
ID:dvi_21042 |
Coordinate:scaffold_13049:22881378-22881528 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_11376:1]; CDS [Dvir\GJ11329-cds]; intron [Dvir\GJ11329-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAAAGAATCCAAACATGCTTGCATCAGTTACGAGGATGCAGTCAAACGTGGTAAGTGTTAACCTTCAACACCTGAGTACAACACTGCCAGTTCTGCAGACCACAAAGGAATGAAGGGAAAAATGGGTCATACGAGTACACATAAGGTTCTAGAACAAAAACAATAATAGTATGCACCAACACAGGTGTAATTGCGCTTTATCGATTCTCTTATTGTTTCCACTCTATTTCCATGCTTTTTTTATTGTTGCC **************************************************....(((((..((((((...(((..((......((((.......))))...))..)))..))))))........((((((((.............(((....))).............))))).))))))))((((((....))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060683 160_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
V047 embryo |
SRR060654 160x9_ovaries_total |
V053 head |
SRR060663 160_0-2h_embryos_total |
GSM1528803 follicle cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................GTTTCCACTCTATTTCCATGCT............... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ACAAAAGGTTCTAGGACAA.............................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................CAGTTACGAGGATGCAGTC................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AAAGAATCCAAACATGCTTGCAT................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CAAACATGCTTGCATCAGTTACGAGGAT...................................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................CAGTTACGAGGATGCAGT................................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................AGTTACGAGGATGCAGTA................................................................................................................................................................................................................ | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................ACACAGCCAGCTCTGCAGAGC...................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TCCTAGTGTTTCCATTCTATT....................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................TCATACGAATACACATGAGA......................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................AAGAGAATACAGTCAAACGT.......................................................................................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................GGTACGTGTTAAACTTCGA....................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CTCTTATTGTTTCCAT............................. | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................AGGCTGTATTCAAACGTG......................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................CGTAAGGGTTAAACTTCAA....................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TTTTCTTAGGTTTGTACGAACGTAGTCAATGCTCCTACGTCAGTTTGCACCATTCACAATTGGAAGTTGTGGACTCATGTTGTGACGGTCAAGACGTCTGGTGTTTCCTTACTTCCCTTTTTACCCAGTATGCTCATGTGTATTCCAAGATCTTGTTTTTGTTATTATCATACGTGGTTGTGTCCACATTAACGCGAAATAGCTAAGAGAATAACAAAGGTGAGATAAAGGTACGAAAAAAATAACAACGG
**************************************************....(((((..((((((...(((..((......((((.......))))...))..)))..))))))........((((((((.............(((....))).............))))).))))))))((((((....))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
GSM1528803 follicle cells |
SRR060670 9_testes_total |
M047 female body |
V116 male body |
M028 head |
SRR060664 9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................CCCTTTTTACGCAGTAAGCT..................................................................................................................... | 20 | 2 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................CGGTCAAGACGTCTGGTGTT.................................................................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................CGTCAGTTTGCACCATTCAC.................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGTGTCATGTTGTGACGGT.................................................................................................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CTACGTCAGTTTGCACCA....................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGTGGGCTCTTGGTGTGACGGT.................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TTGTCTCATGTTGTGACGGT.................................................................................................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGTCCCATGGTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 4 | 0.75 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGTCTCTTGTTGTGACGGT.................................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AGGGGTTTCCTTAGTTCCCT..................................................................................................................................... | 20 | 3 | 5 | 0.40 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GGGGTTTCCTTAGTTCCCTA.................................................................................................................................... | 20 | 3 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................TGGTCTCATGGTGTGAGGGT.................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGTCACATGGTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGGAAGTGGTGGGCGCATG............................................................................................................................................................................ | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................TGGGCGCTTGTTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGTTCTCATGGTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGTCTAATGCTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................GCTGTCAATGCTACTACGTC.................................................................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGGCTAATTTTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGACGCCTGTTGAGACGGT.................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................TGGACACTTGGTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AGGGGTTTCCTTAGTTCCC...................................................................................................................................... | 19 | 3 | 16 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ....................................................................TTGGTCTCTTGTTGTGACGGT.................................................................................................................................................................. | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGCCGCTTGTTGTGACGGT.................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................GGGTTTCCTTAGTTCCC...................................................................................................................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................CGGTCAAGACGACTGGTTTTT................................................................................................................................................. | 21 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................GTCCTCATGGTGTGACGGT.................................................................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCAAGACGCATGGTTTTTCC............................................................................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................GTCAAGACGACTGGTTTTT................................................................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:22881328-22881578 - | dvi_21042 | -------------AAAAGAATCCAAACATGCT---TGCATCAGTTACGAGGATGCAGTCAAACGTGGTAAGTG----------TTAA--C--CTTCAACACC-----------------------TGAGTA------------------------------------------------CAACAC-------------TGCCAGTTCTGCAGACCACAAAGGAATGAAG-GGAAAAATGGGTCAT---A-----------------------CG-----AGTAC---ACATAAG-GTTCTA--------------------------------G---------AACAA------AAACAATA---ATAG-----------------------------------TAT--------GCACCAA---CACAGGTGTAATTGCGCT-TTATCGA------TTC-TCTTA--TTG-----TTTCCA-----------------------------------CT---------CTATTT-CCATGCTTTTTTTAT-------------TG-----------------------------------------TTG-CC |
| droMoj3 | scaffold_6680:17496222-17496465 - | -------------AAAAGAATCTAAACATGCT---TGCATCAGTTACGAGGATGCAGTCAAACGTGGTAAGTG----------TTAA--C--CTCGAATACACATAAGGACAGGTGCTATC---------------------------------------------------------------------------TGAATTGC------AGACAACAAAAC----ATG-GGGAAAATGGGTCATTCAA-----------------------GT-----ACTTA--------AG-GCTCTA--------------------------------G---------AACAA------AAACAATA---ATAA-----------------------------------TGG--------ACACCAA---TACAGTTATAATTACAC---TATCGA------TTC-CATTG--TGGT-----TTCCA-----------------------------------CT---------ATACTT-CCATGCTTTT---AT-------------TG-----------------------------------------TTGCAC | |
| droGri2 | scaffold_15110:18155608-18155830 - | -------------AAAAGAATCCAAACACGCT---TGCATCAGTTATGAGGAGGCAGTCAAACGTGGTAAGTG----------CTAA--C--CTTCAAA-----------------------------------TCC-CA----------------------------------CAGCAAGG-------------------------------CAACA--------AAA-ACGAAAATGGGTCAT--TCGA----------------GTGTACA-------------ACATAAG-GTTCTA--------------------------------G---------AAGAAAAA------CAATA---ATAG-----------------------------------T-----ATAATGCACCAA---CACAGGTGTAATTGCGCT-TTATCGA------TTC-TGTTG--TTG-----TTTTCA-----------------------------------TT---------CTATTCCCCATGCTTT----------------------------------------------------------------T-AT | |
| droWil2 | scf2_1100000004540:539829-540150 - | -------------AAAAGAGTCTAAACACGCC---TGCATAAGTTACGAGGATGCAGTCAAACGTGGTAAGTAACCACTTGGGTTAA--C--CTTCATTACA------------------------------------------------------T-------------TG---------------------------GT--C---------------------------ACAAAATGGGTCAT--CACGGTTCTCTGCGGCCGTCGTCTACG-------------ACGTCAGAGTTCTAGAATCCGCAGCAAGAAG-G-CACTAGTACCAGCAGCACAGGGCAAAGCAACAAAAACAATT---ATACATGTGTGCACGCGTGCGTTTGTCTGTGTGTGTGTGT--------GCATACCAA---TACAGGTGTAA-TGCGCT-TTATCGA------TTC-TATTGTGTTCTTA--TTCCCA-----------------------------------AT---------CTATTT-CCATTTCCAT---GC------------GTG-----------------------------------------T----T | |
| dp5 | XR_group6:11827336-11827599 - | dps_3161 | -------------AAAGGAGTCGAAACATGCC---TGCATCAGCTACGAGGATGCAGTCAAACGTAGTAAGTGA---------TTAA--C--CTTCAAAGCCTGTTAGTGTTGGCTCCATC---------------------------------AAT-------------TGACCAGCACAGCACCTTCCGCGCTGGCCGCCAC---------------------------TTAAAATGGGTCAC--TA-----------------------CGACACGAATCG---ACGTCA--GTTCTGGAAGCC--------------------------T---------AAAAACAA---AAACAATT---CTAC-----------------------------------T--------GCCCACCAA---CACAGGTGTAACTGCGCC-TTATCGA------TTC-TATTG--TTG---TGCTCCCA-----------------------------------TT---------CTACCT-CCATG-----------------------------------------------------------------------C |
| droPer2 | scaffold_20:1748961-1749224 - | -------------AAAGGAGTCGAAACATGCC---TGCATCAGCTACGAGGATGCAGTCAAACGTAGTAAGTGA---------TTAA--C--CTTCAAAGCCTGTTAGTGTTGGCTCCATC---------------------------------AAT-------------TGACCAGCACAGCACCTTCCGCGCTGGCCGCCAC---------------------------TTAAAATGGGTCAC--TA-----------------------CGACACGAATCG---ACGTCA--GTTCTGGAAGCC--------------------------T---------AAAAACAA---AAACAATT---CTAC-----------------------------------T--------GCCCACCAA---CACAGGTGTAACTGCGCC-TTATCGA------TTC-TATTG--TTG---TGCTCCCA-----------------------------------TT---------CTACCT-CCATG-----------------------------------------------------------------------C | |
| droAna3 | scaffold_13337:19407393-19407666 - | -------------CAAGGAGTCCAAGCACGCT---TGCATCAGCTACGAGGATGCCGTTAAGCGAGGTGAGC------------TCTCC---------------------------------GGACGGAGACCAAGTGTC-GA-TGGA-AGTGAAAA-------------TGAC---------------------------CG----------------------------TGGAAATGGGTCAC--TG-----------------------CT-----AATCA---ACGCAA--CTT---GACCAA--------------------------T---------AAAAACAA---------CA---ACAA-----------------------------------TGCT-CACAGAACACCAA---CACAGGTGTGATTGCGCT-TTATCGA------TTC-CATTG--TTG---TGCTTCCACAAATTCTAATTGACACCAGTG-------GCCACCACTCGTTCCAACTACTC-CCAAACCCCT---GC-------------CG-----------------------------------------T----T | |
| droBip1 | scf7180000396360:602184-602411 - | -------------CAAGGAGTCCAAGCACGCT---TGCATCAGCTACGAGGATGCCGTTAAGCGAGGTGAGC------------TCTCC---------------------------------GGACGGGCA---ACTGTC-GA-TGGA-AGAGAAAA-------------TGAC---------------------------CG----------------------------TGGAAATGGGTCAC--TG-----------------------CT-----AATCA---ACGCAA--CTT---GACCAA--------------------------T---------AAAAACAA---------CA---ACAA-----------------------------------TGGGCCCCAGAACACCAA---CACAGGTGTGATTGCGCT-TTATCGA------TTC-CATTG--TTG---TGCTTCCACAAATTCTAAT----------------------------------------------------------------------TG-----------------------------------------T-C-AC | |
| droKik1 | scf7180000302391:632268-632535 - | AAAAGATGGGCACCAAGGAGTCCAAGCACTCAAACAGCATCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTG----------TGCTGC----CC------------------------CCTGGCTGGACC--AAA----------------------TTCCC-----------TGG----------------------GA--------------------------AA-GTGAAAATGGGTCACACAG-----------------------CT-----AATCAACGACGTCG--CTTCTA-------------GAAA-A-CAGAA----------------------CAA---------CACC---AA-----------------------------------CAT---AC---------AT---ACAGGTGTGATTGCGCT-TTATCGA------TTCCCATTG--TTGTTGTGCTCTCACGTTTGCTGAC------CACTACACCCTGGACACAAG---------GTGG---------TT---CCAC-------------TG-------------------------------------------C-CC | |
| droFic1 | scf7180000454113:2454503-2454751 - | -------------CAAGGAGTCGAAACACTCCAATAGCATCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTT----------TTAA--CTGCCTCAA--------------------------------A---TTT-TG-GA-TGGTA--------ACACCCTCCGG--T---------------------------GACTGC---------------------------GGAAAATGGGTCACTCTG-----------------------CT-----AATCA-----------------------------AGAAA-A-CAGAACCCCC-GC---------AATAGCAA---------CAGCAACAA-----------------------------------C----------------AAACCGCAGGTGTGATTGCGCTGCAATCGA------TTC-CATTG--TTGTTG--CTTCCACGATTTCTGCC------CAGTG-------G-----------------------------TTCCTATAC-------------TGACCA-----------------------------------------CC | |
| droEle1 | scf7180000491193:4130361-4130565 + | -------------CAAGGAGTCCAAGCACTCCAATAGCATCAGCTATGAGGATTCGGTGAAGCGCGGTGAGTGAG--------TCCTCC---------------------------------GGATGGGTA---TTT-TG-GA-TAGTA--------ACACCC-CCGGTG-----------------------------GCC-------------------------AG-TGGAAAATGGAGTAC--TA-----------------------CT-----AATCA-----------------------------AGCAA-A-CAAAATCCCC--------------------------------------------------------------------------------------------------ACT-----------GCAATCGA------TTC-CATTG--TTGTTG--CTCATACGATTTCTGAC------CAGTG-------C-----------------------------CTCCTCCAC-------------TG----------------------------------------------- | |
| droRho1 | scf7180000773422:74500-74711 - | -------------CAAGGAGTCCAAACACTCCAATAGCATAAGCTACGAGGATTCGGTAAAGCGCGGTAAGTGAA--------TTCTCC---------------------------------GGATGGGTA---TTT-TG-GGTTGGTA--------ATATAC-CCGGTG-----------------------------GCG-------------------------AG-TGGAAAATGGGTTAC--GG-----------------------CT-----AATCA-----------------------------AGAAA-A-CAGAAACCCC--------------------------------------------------------------------------------------------------GCT-----------GCAATCGA------TTC-CATTG--TTATTG--GTCGCACCATTTCTGAC------GAAAG-------T-----------------------------TTTCCCCAC-------------TGACCA-----------------------------------------CC | |
| droBia1 | scf7180000302193:5818404-5818684 - | -------------CAAGGAGTCCAAGCACTCCAATAGCATCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTG----------TCCTCC---------------------------------GCTCGG-AA---TTT-TC-GTGTGCCA--------GTGCCC-CCCGTC----------------------------GGA--------------------------AG-TGGAAAATGGGTCAC--TG-----------------------CT-----AATCA-----------------------------AGAAACA-CAGAGAACCCCGC---------ACAAGCAA---------CAAC----------------------------------------------AC---------GG---GCAGGT-----------GCAATCGA------TTC-CATTG--TTGTTG--CGCCCACGACTTCTGAC------CAGTG-------G-----------------------------CTCCCGCACCACCCCCGCT-CCTGACCACCACCCTCTGCTC---CTCCACAACCTGCAATTGAAATTC-CC | |
| droTak1 | scf7180000415254:716685-716927 - | -------------CAAGGAGTCCAAACACTCCAATAGCATCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTG----------TCCTCC----C----------------------------AATTGG-AA---TTT-TGT--------------------------------------------------------------------------------------AG-TGGAAAATGGGTCAC--TG-----------------------CT-----AATCA-----------------------------CGAAA-AGCAGAACCCCC-GC---------AGAAGCAA---------CACCAACAA-----------------------------------C-----AC---------AG---GCAGGT-----------GCAATCGATTCCGATTC-CATTG--TTGTTG--CG-------CTTCTGAC------CAGCG-------G-----------------------------TTCCCCCGC---------CCCCTGACCCA------CTGCTC---CTACTGATCCTGG-------ATTT-CC | |
| droEug1 | scf7180000409466:5084005-5084247 - | -------------CAAGGAGTCCAAGCACTCCAATAGCATCAGCTATGAGGATTCAGTGAAGCGCGGTGAGTG----------TCCACG---------------------------------GGCTCGAAA---TTT-TT-GATTGTT---------ATTCCCTGCTGTA-----------------------------GGC-------------------------ATTGGGAAAATGGGTAAC--TG-----------------------CT-----AATCA-----------------------------AGAAA-AAGAGAACCCCC--G---------AAAAGCAA---------CAGCAACAA-----------------------------------C-----AA---------AA---GCAGGT-----------GCAATCGA------TTC-AATTT--GTG--------------------TC------CAGTG-------A-----------------------------TTATTCCAC------------CTGACCAACACCATCTTCTCTCTCTC------------------TTT-CC | |
| dm3 | chr3L:19864502-19864717 + | -------------CAAGGAGTCCAAGCACTCCAATAGCGTCAGCTACGAGGATTCGGTGAAACGCGGTGAGTC----------TCCTCC---------------------------------GGCTGGTTA---CTTG--T---G------------GGG-------------C----------------CGTCTCCCAGCCGC---------------------------AGAAAATAGGTCAC--TG-----------------------CC-----AATCA-----------------------------AGAAA-A-CAGAACCCCCTGC---------GAAAGCAA---------CAAC---AA-----------------------------------CGT---AC---------AA---GCAGGT-----------GCAATCGA------TTC-CATTGTGTTGTTG--CGCCCACGTCTTCTGAC------CAACG-------G---------------------------------------------------------------------------------------------------G | |
| droSim2 | 3l:19458456-19458671 + | dsi_12627 | -------------CAAGGAGTCCAAGCACTCCAATAGCGTCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTC----------TCCTCC---------------------------------GGCTGGTTA---CTTG--T---G------------GGG-------------C----------------CGTCTCCCAGCCGC---------------------------TGAAAATAGGTCAC--TG-----------------------CT-----AATCA-----------------------------AGAAA-A-CAGAACCCCCTGC---------GAAAGCAA---------CAAC---AA-----------------------------------CGT---AC---------AA---GCAGGT-----------GCAATCGA------TTC-CATTGTGTTGTTG--CGCCCACGTCTTCTGAC------CAACG-------G---------------------------------------------------------------------------------------------------G |
| droSec2 | scaffold_29:509932-510147 + | -------------CAAGGAGTCCAAGCACTCCAATAGCGTCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTC----------TCCTCC---------------------------------GGCTGGTTA---CTTG--T---G------------GGG-------------C----------------CGTCTCCCAGCCGC---------------------------AGAAAATAGGTCAC--TG-----------------------CT-----AATCA-----------------------------AGAAA-A-CAGAACCCCCTGC---------GAGAGCAA---------CAAC---AA-----------------------------------CGT---AC---------AA---GCAGGT-----------GCAATCGA------TTC-CATTGTGTTGTTG--CGCCCACGTCTTCTGAC------CAACG-------G---------------------------------------------------------------------------------------------------G | |
| droYak3 | 3L:20716955-20717183 - | -------------CAAGGAGTCCAAGCACTCCAATAGCGTCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTC----------TCCTCC---------------------------------GGCTGGTTA---CTAG--TACTT------------GTA-------------C----------------CGTCTCACAGCCGC---------------------------AGAAAATAGGTCAC--TG-----------------------CT-----AATCA-----------------------------AGAAA-A-CAGAACCCCCTGC---------AACAGCAA---------CAACCACAA-----------------------------------CTT---TC---------AA---GCAGGT-----------GCAATCGA------TTC-CATTGTGTTGTTG--CGCCCAAGTCTGCAGAC------CAAAT------AT-----------------------------AT---------------------------------------------------------------TTT-CC | |
| droEre2 | scaffold_4784:19602693-19602935 + | -------------CAAGGAGTCCAAACACTCCAATAGCGTCAGCTACGAGGATTCGGTGAAGCGCGGTGAGTC----------TCCTCC---------------------------------GGCTGGCTA---CTTG--TACTT------------GTA-------------C----------------CGTCGTACAGCCGC---------------------------AGAAAATAGGTCAC--TG-----------------------CT-----AATCA-----------------------------AGAAA-A-CAGAACCCCCTCC---------AAAAGCAA---------CAACAACAA-----------------------------------CTT---AC---------AA---GCAGGT-----------GCAATCGA------TTC-CATTGTGTTGCTG--CGCCCACGTCTGCTGAC------CAACT------GG-----------------------------ATTCTCCAC-------------TGACCAC------------------------------------CAG-CC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/21/2015 at 04:19 PM