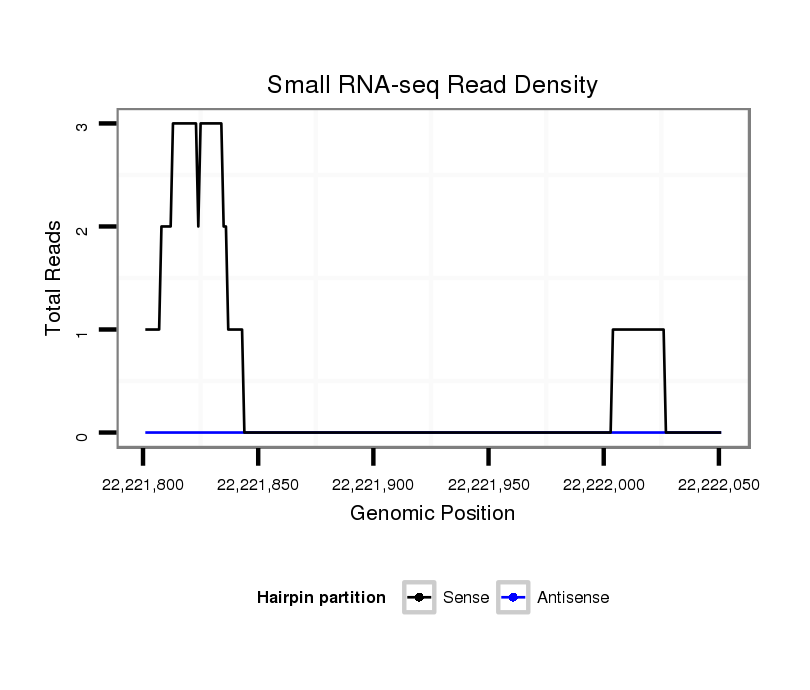
| AACAACAGCAGCAACA----------------------------------ACAACAGCAGCAG---CAACA------------------------------------------------------------------------------------------ACAGCAA--------------------------------------------------------------------------CA-----------------GCAACAACAACAG--------------------------------------------------------------------------------------CAGCA-------------ACAACAGCAACAGCAACAAC---AGCAGCAGCAGCAGCAACTGCAACAACAGCAG----------------------------------------------CAGCAACA--GCAACAACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAACAGCAACAACAGCA-----------------------------------------------------------------------A------------------------------CAA | Size | Hit Count | Total Norm | Total | M020
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| .......GCAGCAACA----------------------------------ACAACAGCA.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ..............................................................................................................................................................................................................................................................................ACAG--------------------------------------------------------------------------------------CAGCA-------------ACAACAGCAACAGC................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................CAACAACAACAG--------------------------------------------------------------------------------------CAGCA-------------ACAACA......................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..................................................ACAACAGCAGCAG---CAACA------------------------------------------------------------------------------------------AC...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| ......................................................CAGCAGCAG---CAACA------------------------------------------------------------------------------------------ACAGCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...................................................................................................................................................................AGCAA--------------------------------------------------------------------------CA-----------------GCAACAACAACAG--------------------------------------------------------------------------------------CAGCA-------------............................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCAGCAACAGCAACAACAGCA-----------------------------------------------------------------------A------------------------------... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAACA..................................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAACAGCAACAA.............................................................................................................. | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................AGCAGCAGCAGCAGCAACTGCA.................................................................................................................................................................................................................................................................................................................................................. | 22 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAACAGC................................................................................................................... | 23 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAAC...................................................................................................................... | 24 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAACA..................................................................................................................... | 25 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................GCAA--------------------------------------------------------------------------CA-----------------GCAACAACAACAG--------------------------------------------------------------------------------------C................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAGCAG----------------------------------------------CAGCAACA--GC.............................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGC......................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................AACAGCAG----------------------------------------------CAGCAACA--GC.............................................................................................................................................................................................................................................................................. | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......AGCAGCAACA----------------------------------ACAACAGCAG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAGCAG----------------------------------------------CAGCAACA--GC.............................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACA--GCAACAACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGC............................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAAC...................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..CAACAGCAGCAACA----------------------------------ACAACAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....ACAGCAGCAACA----------------------------------ACAACAGCA.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAT--------------------------------------------------------------------------------------------------------------------------------CAGCAGCAAC...................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................AA--------------------------------------------------------------------------CA-----------------GCAACAACAACAG--------------------------------------------------------------------------------------CAGCA-------------............................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................AACAGCAACAAC---AGCAGCAGCA.............................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................CAACAGCAACAGCAACAAC---AGCAGCAGCA.............................................................................................................................................................................................................................................................................................................................................................. | 29 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................CAACAGCAACAAC---AGCAGCAGC............................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................AA--------------------------------------------------------------------------CA-----------------GCAACAACAACAG--------------------------------------------------------------------------------------CAGCA-------------ACA............................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................CAACAGCAACAAC---AGCAGCAGCAGCAG.......................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................AACAACAG--------------------------------------------------------------------------------------CAGCA-------------ACAACAGC....................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................CAA--------------------------------------------------------------------------CA-----------------GCAACAACAACAG--------------------------------------------------------------------------------------CA............................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............A----------------------------------ACAACAGCAGCAG---CAACA------------------------------------------------------------------------------------------AC...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............A----------------------------------ACAACAGCAGCAG---CAACA------------------------------------------------------------------------------------------A....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................AACAGCAGCAG---CAACA------------------------------------------------------------------------------------------ACAGCAA--------------------------------------------------------------------------CA-----------------.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |