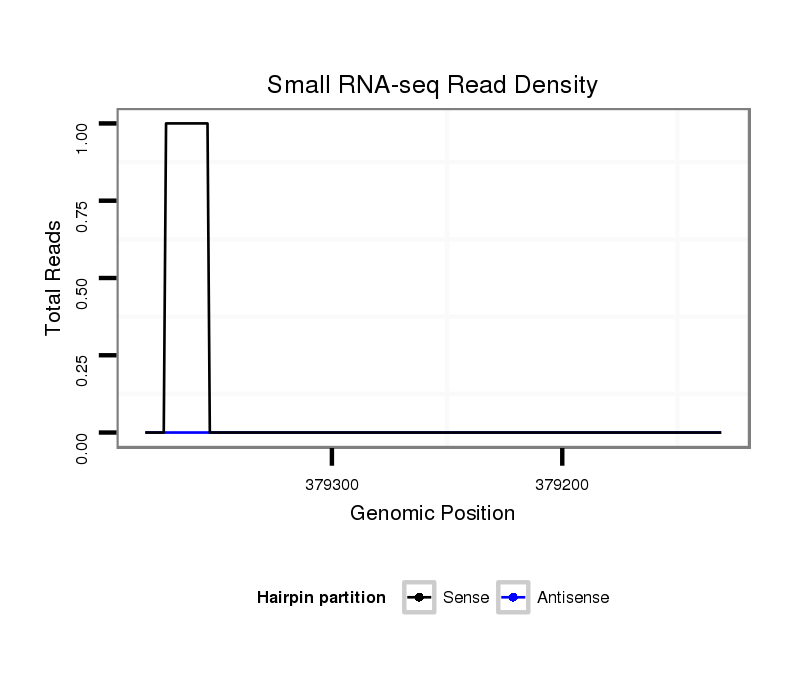
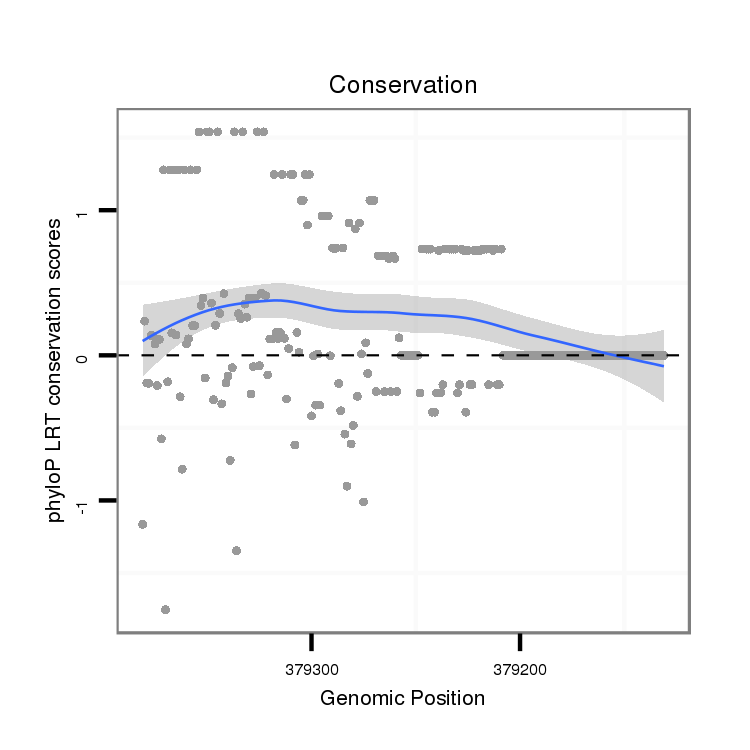
ID:dvi_2080 |
Coordinate:scaffold_12728:379181-379331 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ15411-cds]; exon [dvir_GLEANR_15755:1]; intron [Dvir\GJ15411-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ----------------##################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTACTGACAAACAAAAATGATTGTGGATTTTATTGCTGGTCTCTTTGGAGGTAATATATATTTTTTTTACATTAACTTAATATTTATAATATGTGCAGTAGTGCAGTCATTTTTTTCTCGACAAAGTGCTTCCATTTTACAACTATTAAAAAGTGTCAAGCCCAAAGAGTGGTAAGTCGGCCAAGGTTTATTTTTGTACATTTTTTTTCCGGAACAATTTTTTTATGTGTAATTGGTTTGAAAGTTAGTAC **************************************************........................................((((((((.((((..(((...........))).....((((.((((((...........))))))..))))........((((......)))).....)))).))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060667 160_females_carcasses_total |
SRR060673 9_ovaries_total |
SRR060677 Argx9_ovaries_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................TAAAGTGCTTGCATTTTAAAAC............................................................................................................ | 22 | 3 | 6 | 3.83 | 23 | 23 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AAAGTGCTTGCATTTTACA.............................................................................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........AACAAAAATGATTGTGGAT............................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................AAGTGCTTGCATTGTACAAC............................................................................................................ | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................AAGTGCTTGCATTTTACAAC............................................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................AAGTGCTTGCATTTTAAAAC............................................................................................................ | 20 | 2 | 6 | 0.50 | 3 | 3 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TATTTTTGGACTTTTTTTTTCC......................................... | 22 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................ATTGTAGGTTGTATTGCTGGTCT................................................................................................................................................................................................................. | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TGAGTGTTATTGGTTTGAAA........ | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............CAGATGATTGTGGGTTTTA........................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
GATGACTGTTTGTTTTTACTAACACCTAAAATAACGACCAGAGAAACCTCCATTATATATAAAAAAAATGTAATTGAATTATAAATATTATACACGTCATCACGTCAGTAAAAAAAGAGCTGTTTCACGAAGGTAAAATGTTGATAATTTTTCACAGTTCGGGTTTCTCACCATTCAGCCGGTTCCAAATAAAAACATGTAAAAAAAAGGCCTTGTTAAAAAAATACACATTAACCAAACTTTCAATCATG
**************************************************........................................((((((((.((((..(((...........))).....((((.((((((...........))))))..))))........((((......)))).....)))).))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060685 9xArg_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060688 160_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................AAAAAAAATGTAATTGAAATCT......................................................................................................................................................................... | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................ATAAAAAAAAAATGTGAATGAATT........................................................................................................................................................................... | 24 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................AAAAAATGTAATTGAAAT........................................................................................................................................................................... | 18 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................ACCACGTAGCCGGTTCCAA............................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TTAAAGAAATACGCATTAA................. | 19 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................AAAAAAATGTGAATGAATT........................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................AAAAAAACGGAATTGAATT........................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................................................................AAAAAAAACACCTCGTTAAAAAA............................ | 23 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12728:379131-379381 - | dvi_2080 | CTACTGA-CAAACAAAAATGATTGTGGATTTTATTGCTGGTCTCTTTGGAGGTAATAT-ATA----------TTTTTTTTACATTAACTTAAT--------------------------------ATTTATAATATGTGCAGTAGTGCA----------------GTCATTTTTTTCTCGACAAAGTGCTTCCATTTTACAACTATTAAAAAGTGTCA---AGCCCAAAGAGTGGTAAGTCGGCCAAGGTTTATTTTTGTACATTTTTTTTCCGGAACAATTTTTTTATGTGTAATTGGTTTGAAAGTTAGTAC |
| droMoj3 | scaffold_6500:30404397-30404496 + | CTGCTGA-CATAGAAAAATGATTGTGGATTTCATTGCTGGCTTACTCGGAGGTAATAATTTAATTTATCTTACCCTTTTTATATTGA---AAT--------------------------------TTCTGTAATA------------------------------------------------T---------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15126:7661605-7661665 + | GTGCTGA-TAAAAAACAATGCTTGTGGATTTTATTGCTGGTCTTTTTGGAGGTAATAT-ATG----------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000002854:604-690 - | GA--------------------------TGATATTCTTGATCAATTGGGAAGAGCAAA-ATA----------TTTTTCTTGCCTTGACTTAAT------------------------------------------------------------GTCAGGATTTCATCTTTTTCTTTTTCAACGA---------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779504:64190-64356 - | TAATTGC--GAACTAAAATTTTGATAAATTTAATTGTTTATGCCATCTCAGGTTGTAT-ATA----------TTTTATATATATTTTTTTA-TAACTGTTATATACAT----------------------------------AAGCGCATGCTGTCATTAATACATGTTTTT---------------------TTTTTAACTCAGTTAAAATATGGCGCACGGCCCAAAAAGTAAT------------------------------------------------------------------------------ | |
| droTak1 | scf7180000412669:8074-8182 + | CAAACAAATATAAGAAATTCATTGTAAATTTTATATTTGCTCTCATAGGAGCTGGTAT-ATA----------C---------------------------------ATTTAAATTTAAATTGGTCAATCATAATTTGTAAACCATTGTA----------------TTT---------------A---------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/19/2015 at 11:32 AM