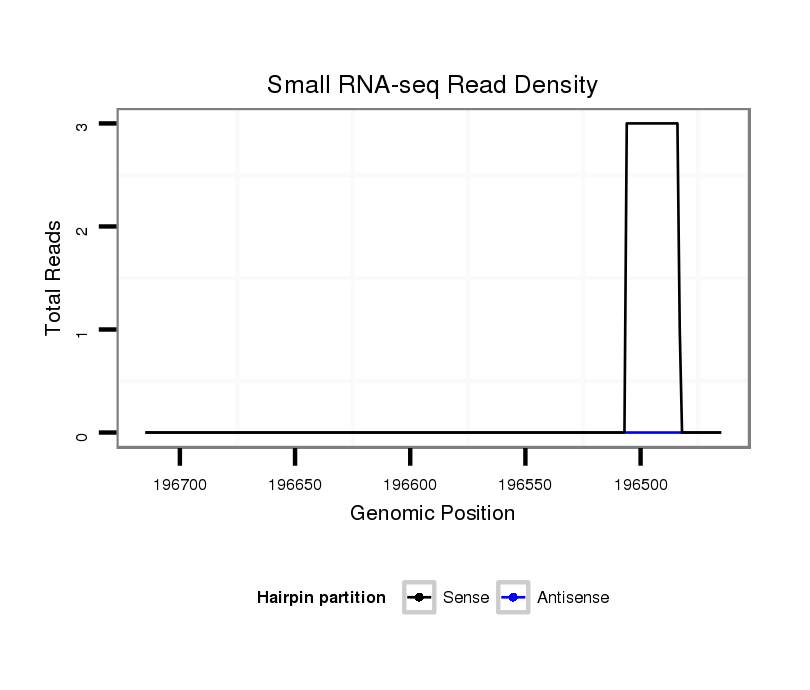
ID:dvi_2075 |
Coordinate:scaffold_12728:196515-196665 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_15766:3]; CDS [Dvir\GJ15415-cds]; intron [Dvir\GJ15415-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGCGACGCCACGGCCCGATAGCGCGAGTGGTGCGAGGCTACAACCTGGCAGTGAGTTTGGTGCGTGGCCACGGCCCGGTAGTGCAGTTGGTGCGAGGTTACGCCCCGATAGTGTAGTTGGAACGTGGCTAGGTCCCGGTAGAGTAGGTACCAGGCTATGGCCCGGTGCAGTAGGTAGTACTAGGCTGCGGCTCAGTAGAGTAGTTGTTGTGAACATAGAGAGAGGGGTATAGTTTGGTCAATATATTCACT **************************************************.(((((((((.((.((((((((..((((...(((((((((.(((......))).)))))..)))).))))..)))))))).)).))))........(((((.(((....))).))))).....(((((.....))))).))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060675 140x9_ovaries_total |
SRR060681 Argx9_testes_total |
M047 female body |
GSM1528803 follicle cells |
SRR060677 Argx9_ovaries_total |
SRR060687 9_0-2h_embryos_total |
V053 head |
M027 male body |
SRR060686 Argx9_0-2h_embryos_total |
V116 male body |
M028 head |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TGAACATAGAGAGAGGGGTATAG................... | 23 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GGCCCCGTAGCGCAGTTGG................................................................................................................................................................. | 19 | 2 | 2 | 1.50 | 3 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GGCCCCGTAGCGCAGTTGGT................................................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGAACATAGAGAGAAGGGTATAGTTC................ | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGAACATAGAGAGAGGGGTATAGT.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGAACATAGAGAGAAGGGTATAGA.................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGAACACAGCGAGAGGGGTCT..................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............CGATAGCACGAGTGGAGCG......................................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GTGCGGCTCAGTAGAATCGTT.............................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................GCCCTGTAGCGCAGTTGGT................................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............CGATAGCACGAGTGGAGCA......................................................................................................................................................................................................................... | 19 | 3 | 8 | 0.25 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GGCCCTGTGGCGCAGTTGGT................................................................................................................................................................ | 20 | 3 | 9 | 0.22 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GATCGCGCAAGCGGTGCGAGG...................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CGAGTGGTCTAAGGCTACA................................................................................................................................................................................................................. | 19 | 3 | 17 | 0.18 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ........................................................................GCCCTGTAGCGCAGTTGG................................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................GTTAGGTCCCGCTAGAGA........................................................................................................... | 18 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| .............................................TGGCAGTGAGATTAGTTCGT.......................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GGCCCCGTAGCGCATTTGGT................................................................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CGAGTGGTCCAAGGCTTCA................................................................................................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................TGGCAGTGTGGTTGGTCCG........................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................CGAGTGGTCTGAGGCTGCA................................................................................................................................................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................TGAAGACAGCGAGAGGGGT....................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
ACGCTGCGGTGCCGGGCTATCGCGCTCACCACGCTCCGATGTTGGACCGTCACTCAAACCACGCACCGGTGCCGGGCCATCACGTCAACCACGCTCCAATGCGGGGCTATCACATCAACCTTGCACCGATCCAGGGCCATCTCATCCATGGTCCGATACCGGGCCACGTCATCCATCATGATCCGACGCCGAGTCATCTCATCAACAACACTTGTATCTCTCTCCCCATATCAAACCAGTTATATAAGTGA
**************************************************.(((((((((.((.((((((((..((((...(((((((((.(((......))).)))))..)))).))))..)))))))).)).))))........(((((.(((....))).))))).....(((((.....))))).))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060682 9x140_0-2h_embryos_total |
M047 female body |
SRR060674 9x140_ovaries_total |
V116 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................GCCGATGTTGGACCGCCACTCT................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TGCCATCTCATCCATTGTGCG................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................AGCACGCTCCGGGGTTGGA............................................................................................................................................................................................................. | 19 | 3 | 8 | 0.38 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| ...................................................................................................................................................................................................ATCTCATCAACAACACTTGTATCT................................ | 24 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CTCATCAACAACACTTGTATCTCTCT............................ | 26 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................CACAACGCTCCGGGGTTGGA............................................................................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................ACCCTTGTATCTCTCTTCCCT....................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................ATCAACCATACATGTATCTCTCT............................ | 23 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................ACAACACTTGTATCTCTCT............................ | 19 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................AAAAACCACTTGTATCTCTCT............................ | 21 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................ACGCTCCGGGGTTGGATC........................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12728:196465-196715 - | dvi_2075 | TGCGACGCCACGGCCCGATAGCGCGAGTGGTGCGAGGCTACAACCTGGCAGTGAGTTTGGTGCGTGGCCACGGCCCGGTAGTGCAGTTGGTGCGAGGTTACGCCCCGATAGTGTAGTTGGAACGTGGCTAGGTCCCGGTAGAGTAGGTACCAGGCTATGGCCCGGTGCAGTAGGTAGTACTAGGCTGCGGCTCAGTAGAGTAGTTGTTGTGAACATAGAGAGAGGGGTATAGTTTGGTCAATATATTCACT |
| droRho1 | scf7180000775161:413-456 - | -----------------------------------------------------------------------------------------------------------------------------GGTTAGGTTAGGTTAGGACGGCTACCAGGCTATAGCCTGGTACA---------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/19/2015 at 11:31 AM