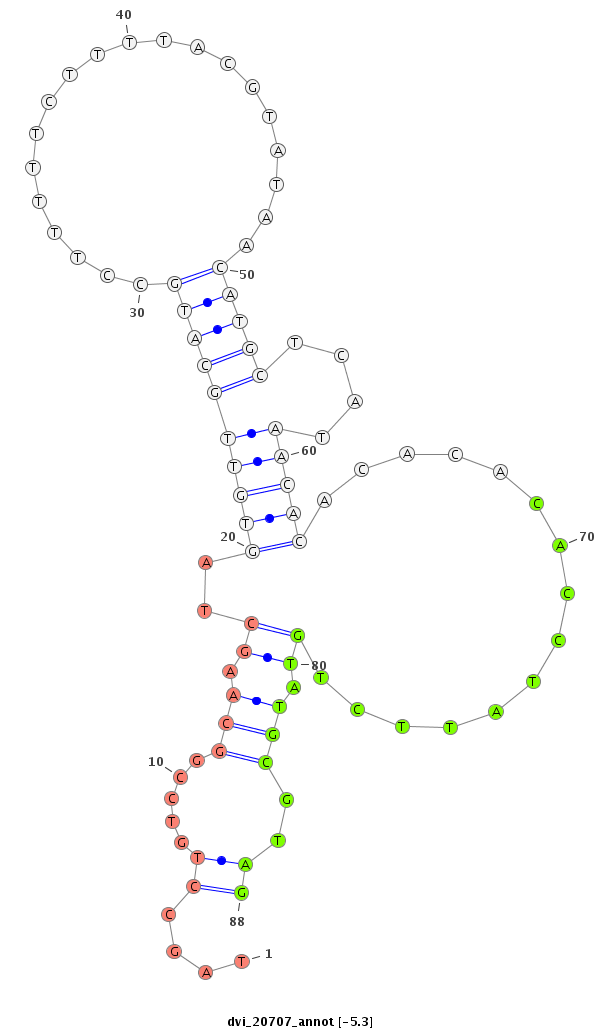
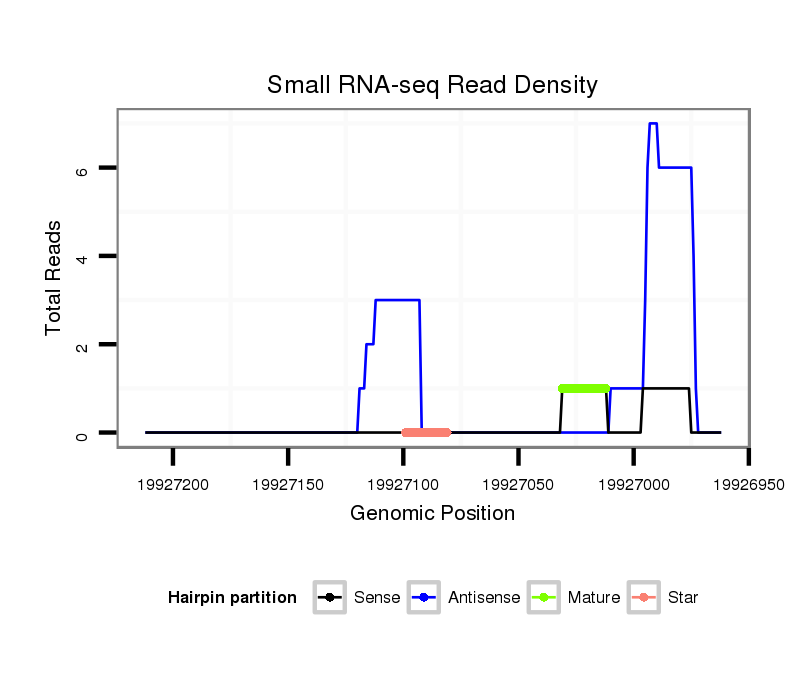
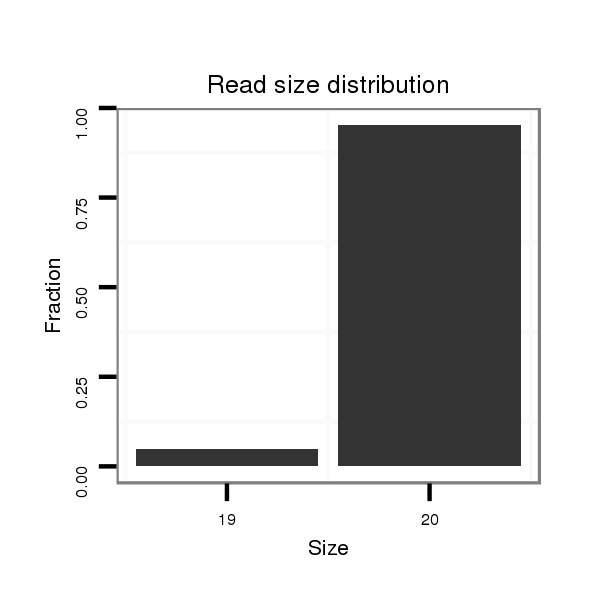
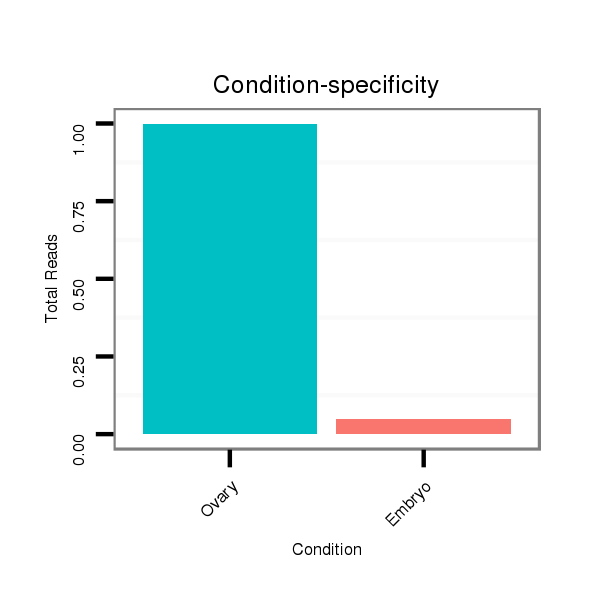
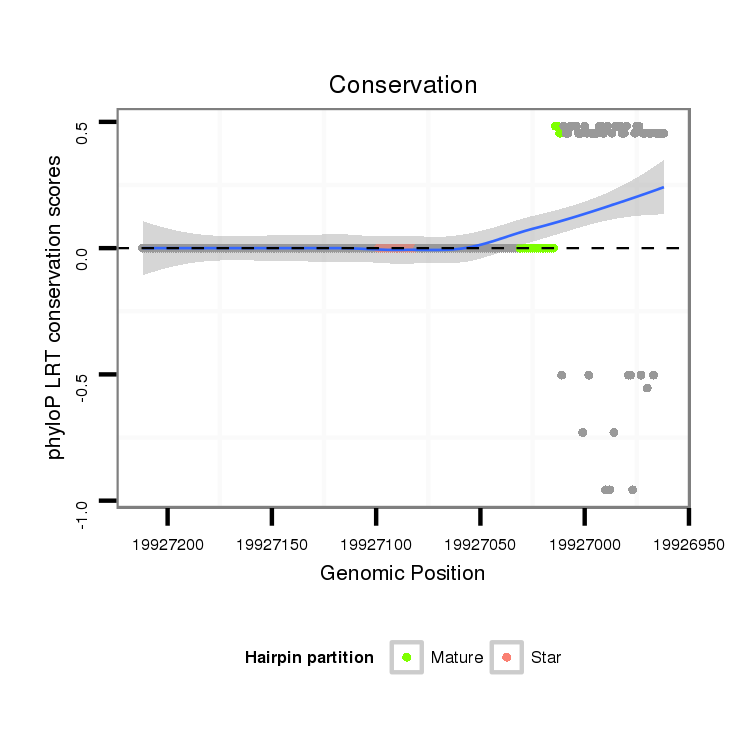
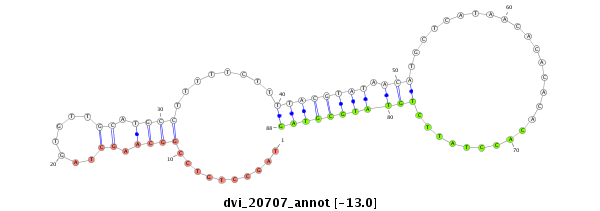
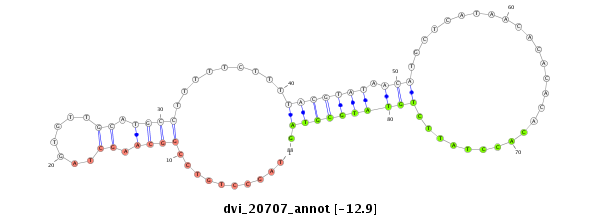
ID:dvi_20707 |
Coordinate:scaffold_13049:19927012-19927162 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.0 | -13.0 | -12.9 | -12.9 |
 |
 |
 |
 |
exon [dvir_GLEANR_11506:6]; CDS [Dvir\GJ11470-cds]; intron [Dvir\GJ11470-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAGTTAGCAAAAAAGGGGGAAAAATGGAATAAAAAACCTATAGAAATTTGAAATAGGCCATAGCATATTAAGCAATAGTGATAGGAAAAAACCGACCCGGCGGGGAATCTTGATAGCCTGTCCGGCAAGCTAGTGTTGCATGCCTTTTTCTTTTACGTATAACATGCTCATAACACACACACACCTATTCTGTATGCGTAGGAGGAATTGGCTCGGCCCAAGGACCTAAAGCTGACCAAACGACGCCGGCG *****************************************************************************************************************....((.....(((.((..((((((((((....................)))))....)))))...............)).)))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060658 140_ovaries_total |
V053 head |
SRR060686 Argx9_0-2h_embryos_total |
M027 male body |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................CCCAAGGACCTAAAGCTGACC.............. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAACACACACGCAGCTATT.............................................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................CACCTATTCTGTATGCGTAG.................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAAGACACACGCAGCTATTC............................................................. | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................TCATAACACACACCCGCCTCT............................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAAGACACACTCTCCTAT............................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAACACACACGCAGCTAT............................................................... | 21 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAATACACACTCAGCTAT............................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GGGGAATCTTGATAGGGT.................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAATACACACACCGCTAT............................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAACACACACACGCCCTT............................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAACACACACGCAGCTAC............................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TCATAAAACACACACGTCTAT............................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................GAAGTTTGATCGCCTGTCC................................................................................................................................ | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................TAGCCTGTCCGGCAAGCTA....................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTCAATCGTTTTTTCCCCCTTTTTACCTTATTTTTTGGATATCTTTAAACTTTATCCGGTATCGTATAATTCGTTATCACTATCCTTTTTTGGCTGGGCCGCCCCTTAGAACTATCGGACAGGCCGTTCGATCACAACGTACGGAAAAAGAAAATGCATATTGTACGAGTATTGTGTGTGTGTGGATAAGACATACGCATCCTCCTTAACCGAGCCGGGTTCCTGGATTTCGACTGGTTTGCTGCGGCCGC
**************************************************....((.....(((.((..((((((((((....................)))))....)))))...............)).)))..))***************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
SRR060655 9x160_testes_total |
SRR060670 9_testes_total |
SRR060678 9x140_testes_total |
SRR060680 9xArg_testes_total |
SRR060689 160x9_testes_total |
SRR060683 160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................GTTCCTGGATTTCGACTGGTT............ | 21 | 0 | 1 | 3.00 | 3 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GGTTCCTGGATTTCGACTGGT............. | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................TTCCTGGATTTCGACTGGTTT........... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................GGCCGCCCCTTAGAACTATCGGAC................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TCCTTAACCGAGCCGGGTTCC............................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................CTGGGCCGCCCCTTAGAACTATCGGAC................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................GCCCCTTAGAACTATCGGAC................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ACGAGCCGGGTTCCTGGATTT..................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......CGTTTTTTCCCCCTTTTTACCTTACTTT......................................................................................................................................................................................................................... | 28 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................AGAAATGGGCATCCTCCTTA........................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................TACCTTATTTTTGGGTTATGT............................................................................................................................................................................................................... | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................CCTTATTTTTTGGTTATG................................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:19926962-19927212 - | dvi_20707 | AAGTTAGCAAAAAAGGGGGAAAAATGGAATAAAAAACCTATAGAAATTTGAAATAGGCCATAGCATATTAAGCAATAGTGATAGGAAAAAACCGACCCGGCGGGGAATCTTGATAGCCTGTCCGGCAAGCTAGTGTTGCATGCCTTTTTCTTTTACGTATAACATGCTCATAACACACACACACCTATTCTGTATGCGTAGGAGGAATTGGCTCGGCCCAAGGACCTAAAGCTGACCAAACGACGCCGGCG |
| droMoj3 | scaffold_6680:7626849-7626901 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGAAGGAATTGGATCAGCCCAAGCAGCGAAAGCTAGGCAAGCGTCGTCGGCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 08:51 PM