

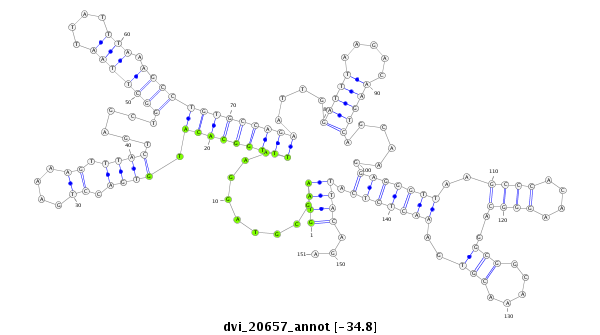
ID:dvi_20657 |
Coordinate:scaffold_13049:19358726-19358876 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
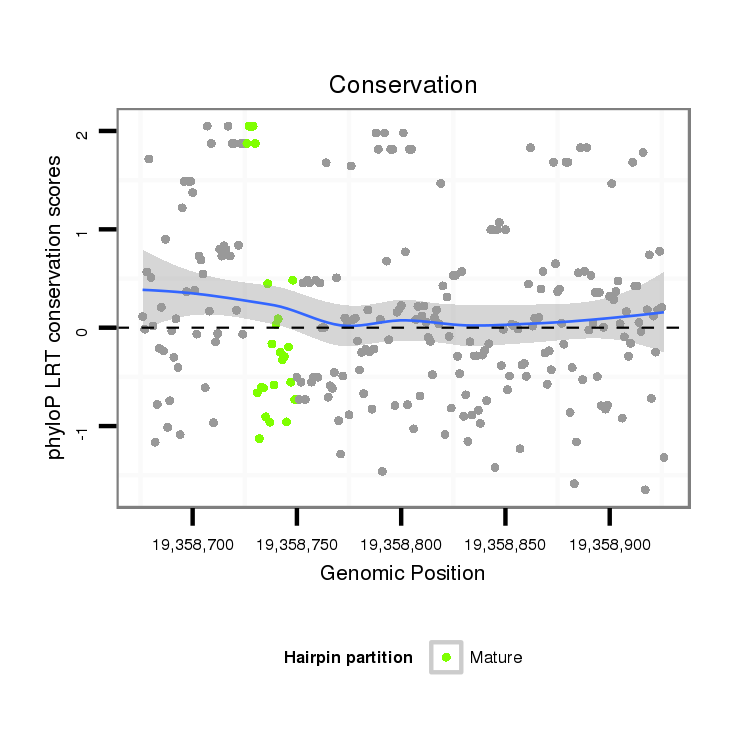
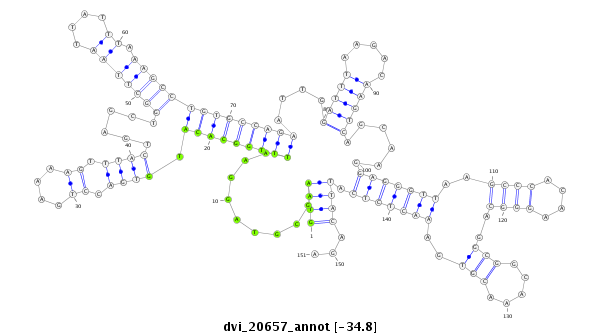
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.8 | -34.8 | -34.8 |
 |
 |
 |
exon [dvir_GLEANR_13684:1]; CDS [Dvir\GJ13823-cds]; intron [Dvir\GJ13823-in]
No Repeatable elements found
| ----------------------############################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATAAAGGCAAAAGATATCCAATATGTTGATCTGGACAAGATTGGCCACGGGTAAGCGTAGGAATTTGGCACATGTGACCTGAAAAGTTTACTGAGCTGGCTTAATTATTTAAAGCCTGTGCCAGAATTGGATTTAAGACAAGTCAGCAAGGAGGGTTAAGCCCACAAGGGCAGGCGGCAAACGTGAAACTCTCATTACAGACGCGCACACTTCATACTGGGCGCATGCTCTCCGCGGATGAAGACGACAAG **************************************************((((.........(((((((((.((((.((....)).))))......(((((.........))))))))))))))....(((((.....)))))......(((((((..((((....))))..(((.....)))..))))))).))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
SRR060681 Argx9_testes_total |
SRR060666 160_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
M047 female body |
SRR060659 Argentina_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGCGTAGGAATTTGGCACATG................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TGATCTGGACAAGATTGGCCACGG......................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGCGTAGGAATTTGGCACCTGT................................................................................................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................GGCGGAACACGTGAAATTC............................................................ | 19 | 3 | 20 | 0.45 | 9 | 0 | 0 | 3 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................ACAAGGGAAGTCGGCAAA...................................................................... | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................ATAAGGGAAGGCGGCAAA...................................................................... | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CCTCTAATTAGAGACGCGC............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................AACTGCACAAGATTCGCCA............................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ACATAAGGGCAGTCGGCAAA...................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TATTTCCGTTTTCTATAGGTTATACAACTAGACCTGTTCTAACCGGTGCCCATTCGCATCCTTAAACCGTGTACACTGGACTTTTCAAATGACTCGACCGAATTAATAAATTTCGGACACGGTCTTAACCTAAATTCTGTTCAGTCGTTCCTCCCAATTCGGGTGTTCCCGTCCGCCGTTTGCACTTTGAGAGTAATGTCTGCGCGTGTGAAGTATGACCCGCGTACGAGAGGCGCCTACTTCTGCTGTTC
**************************************************((((.........(((((((((.((((.((....)).))))......(((((.........))))))))))))))....(((((.....)))))......(((((((..((((....))))..(((.....)))..))))))).))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060671 9x160_males_carcasses_total |
M027 male body |
SRR060672 9x160_females_carcasses_total |
SRR1106727 larvae |
|---|---|---|---|---|---|---|---|---|---|
| .................................................CCATTCGCATCCTTAAAATTT..................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ............................................GGTGCCCTTGCGCAGCCT............................................................................................................................................................................................. | 18 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ....................TATACAAGTAGAGCTGTCC.................................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .......GTTTTCTAAAGGTTAGAC.................................................................................................................................................................................................................................. | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:19358676-19358926 + | dvi_20657 | ATAAAGGCAAAAGATATCCAATATGTTGA---TCTGGACAAGATTGGCCACGGGTAAGCGTAG-GAATTTGGCACATGTGACCTGAAAAGTTTACTGAGC--TG----------------------------GCTTA-------------------------------ATTATTTAAAGCCTGTGCCAGAATTGGATTTAAGACAAGTCAGCAAGG--AGGGTTAAGCCCACAAGGGCAGGCGGCAAAC----------------------------GTGAAACTCTCATTACAGACGCGCACACTTCATACTGGGCGCATGCTC----TCCG-CGGA------TG-AAGACGACAAG |
| droMoj3 | scaffold_6680:7091240-7091468 + | AATAAAGCTTAATTTGTTAAAAATGCAGA---TCTTGTCTAGATTGGCAACGGGTAAGATTTT-AGACTTTTCGCTTTCTTCATGTGAGG--------------------------------------TTAGT-TTAAATT---------------T-----------ATTATTTGCAGCTTGCGCTAAAATCGGCCTTAAGGTAAATCGTCAA-----------------GTTGGCCAACCAGCAAAG----------------------------GCTAACTTTTCATTGCAGAGGCGTTGCCTTTACAGTACACC---GCTG----CTGA-AGGA------TG-ATGACGACAAA | |
| droGri2 | scaffold_15110:17042124-17042362 + | CGAAAATTG-------TTGAAAATGTCGAACTCATGGGCTAGGTTGGCCACGGGTAAGTGTGG-ACAATTAGATT----------------------------A--------TATCTTTTATCTGACATCTAGTTGAG-TT---------------T-----------TATTTTTAAAGCATGTGCAAGAATTGGTTCAAGAACTGGA------GG--CGGCGTAGATTGGCTAGGTCAGCCAATGGCA----------------------------AAAAAATTTCTAATGCAGGCGCGTATGCTGCATACTGCTCCTGTGCTC----TTCA-AGAA------TG-ACGACGATAAA | |
| droWil2 | scf2_1100000004837:905671-905918 + | AAAAAATTAAAACGCAACG-AAATGTCGGTAGCTTGGTTAAGATTGGCCACAGGTAAGAAATA-AAATGAG---------------------TGGAATGG--CT--------ATTCCTTTACATGA--CTTGGGTTAA-TT---------------G-----------TTGGCTAACAGGATACGCCCGTATTGGCTATAAATGGAAT------AATGGAA--ACCATGTGCAAGGACAACCTTCAAAG----------------------------ATGAAGTTTCCTTCGCTGGCTCGCTATCTACATGCCACTTGGATGAAC----CGCA-AGGA------TA-AGAAGTCCGGC | |
| dp5 | XR_group6:1741072-1741316 - | A-------------------------TGGCGTCTTGGGTACGATTGGCCACAGGTAAGAAAGA-AAGC--T---------------------TTTAATAAGTAAGTG----------------------------CTGTAGATTCTGAATGTAATTGGAAA------AACCCTGGGCAGTGTGTGCACGAACCGGCTTTCAAATTGGCCAGCGGAAGGGAG--C-------------------------TCTGGTGCTGGGCCATCCGATGGGCATAGTGAAGTCTCCTCTTCAGGTGCGTCGTCTTCATGTCGCTCGCATGCTCGCCTG-CAA-GGGCAACGACA-AAGATGGCAAA | |
| droPer2 | scaffold_9:33884-34128 - | A-------------------------TGGCGTCTTGGGTACGATTGGCCACAGGTAAGAAAGA-AAGC--T---------------------TTTAATAAGTAAGTG----------------------------CTGTAGATTCTGAATGTAATTGGAAA------AACCCTGGGCAGTGTGTGCACGAACCGGCTTTCAAATTGGCCAGCGGAAGGGAG--C-------------------------TCTGGTGCTGGGCCATCCGATCGGCATAGTGAAGTCTCCTCTTCAGGTGCGTCGTCTTCATGTCGCTCGCATGCTCGCCTG-CAA-GGGCAACGACA-AAGATGGCAAA | |
| droAna3 | scaffold_13337:16527095-16527334 - | AAAAGAAAAAT--------AAAATGTTGGCTTCATGGACAAGATTAGCAACGGGTAAGAGGAG---TT--T---------------------TCTTTTCC--AATTTTAATATAAT------------------TTTAATT---------------TATAAAATTTTAA-ATCACTAAGCTTGCGCCAAAATTGGCCTGAAAGTGGGCCAACCCAACAATG--G-------------------------CCAGGTTCGCAATCAAATGGGGAGTCTTCTGCAGTCTCCACGACATGCCCGGTCTCTGCACGCCTCCT-------TGGGCTT-GTGG-------------GATGACAAG | |
| droBip1 | scf7180000396641:53143-53192 - | AAAAAATTAAG--------AAAATGTTGGCTTCCTGGACAAGATTAGCAACGGGTAAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droKik1 | scf7180000302486:2795649-2795797 - | --------------------------TGGTGACTTGGGGAAGATTGGCCACAGGTAAGCCCATCATGC--T---------------------TACCATTA--TA--------CCTCTTTT----------------------------ATCTAATTGGTGG------AGTCCATGTCAGCCTGTGCCAGAATCGGGCTCCAAATTGGCCACCCGAATGGGG--G------------------------------------------------------------------------------------------------------------------TCACTTCAGAGGCCTGCAGC | |
| droBia1 | scf7180000302428:4030599-4030830 - | CAAAATGC------------------TAGGGACTTTGACTAAAGTGGCCGCGGGTAAGACTAA---AC--T---------------------TATCTTGT--TAGC----------------------------TTAGCTTTTTCT------AAA-AAAAG------AAACCATGGCAGCCTGTGCTAGGATGGGTCTGAGGATTGGCCAGGCTAATGGAG--C----TCATGG------------------GGGACTGACCCAGATGGGTAACTTTGCCCAGCTACCCAAACATGCCCGCTCCTTGCACGC-------ATCCTCATCGCT-TTGGGA------TG-ATAAGGATAAG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
Generated: 05/21/2015 at 04:11 PM