| CAACA-------------------GCAGCAGGAACAGCAGCA---GCAACAATATCAAC------------------AGCAACAC--------------------------------------------------CAGCAGC------AGGAACAGC-----------A---AC---------------------------AA-CAGCAGCAGCAACA---------------------------------------------------------ACAACAA------C----------AGCAGCAGCAACA------------------ATATCAACAGCAACAG----CAACA----------------------ACAGCAGCAGGAACAGCAACAA------------------------------------------------------------------------------------CAGCAGCA-GCA---A------------------------------------------------CAATAT | Size | Hit Count | Total Norm | Total | M020
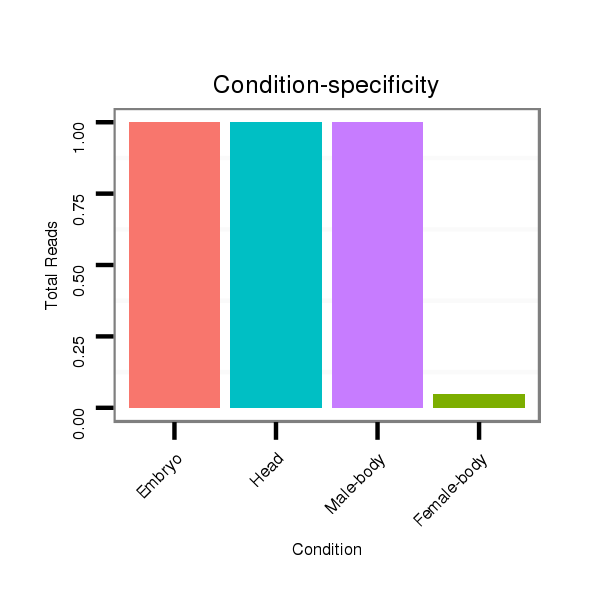
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ..................................................AATATCAAC------------------AGCAACAC--------------------------------------------------CAGCAGC------..................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................TCAAC------------------AGCAACAC--------------------------------------------------CAGCAGC------AGGAAT............................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................AACAATATCAAC------------------AGCAACAC--------------------------------------------------CAGCA............................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................TCAAC------------------AGCAACAC--------------------------------------------------CAGCAGC------AGGAACA.............................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................CA----------------------ACAGCAGCAGGAACAGCAACAA------------------------------------------------------------------------------------C..................................................................... | 25 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------A............................................................................................................................................................................... | 23 | 20 | 0.60 | 12 | 0 | 12 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------................................................................................................................................................................................ | 22 | 20 | 0.45 | 9 | 0 | 9 | 0 | 0 | 0 |
| ............................CAGGAACAGCAGCA---GCAACAA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------ACAGC........................................................................................................................................................................... | 27 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ...................................................................................AC--------------------------------------------------CAGCAGC------AGGAACAGC-----------................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................AACA---------------------------------------------------------ACAACAA------C----------AGCAGCA....................................................................................................................................................................................................................................................... | 19 | 20 | 0.25 | 5 | 0 | 0 | 0 | 5 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................AACA----------------------ACAGCAGCAGGAACAGCAACA........................................................................................................................................................... | 25 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------AC.............................................................................................................................................................................. | 24 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------ACAG............................................................................................................................................................................ | 26 | 20 | 0.20 | 4 | 0 | 3 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................AG----CAACA----------------------ACAGCAGCAGGAACAGCA.............................................................................................................................................................. | 25 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CA---------------------------------------------------------ACAACAA------C----------AGCAGCAGCAACA------------------AT............................................................................................................................................................................................................................. | 25 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................AGCAACAG----CAACA----------------------ACAGCAGCAG...................................................................................................................................................................... | 23 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------ACA............................................................................................................................................................................. | 25 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................A---------------------------------------------------------ACAACAA------C----------AGCAGCAGCAAC.................................................................................................................................................................................................................................................. | 21 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................ACAG----CAACA----------------------ACAGCAGCAGG..................................................................................................................................................................... | 20 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------ACAGCAG......................................................................................................................................................................... | 29 | 19 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................CAGC-----------A---AC---------------------------AA-CAGCAGCAGC................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................................................................................................................................GCAGCAACA---------------------------------------------------------ACAACAA------C----------AGC........................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------ACAGCA.......................................................................................................................................................................... | 28 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................ATCAACAGCAACAG----CAACA----------------------ACAG............................................................................................................................................................................ | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................CAGCAACAA------------------------------------------------------------------------------------CAGCAGCA-GC........................................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .....................................................................................................................................................................................................................CAACA---------------------------------------------------------ACAACAA------C----------AGCAGCAGC..................................................................................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACA------------------ATATCAACAGCAACAG----CAACA----------------------................................................................................................................................................................................ | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................ATCAACAGCAACAG----CAACA----------------------ACAGCA.......................................................................................................................................................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................AGCAGCAGGAACAGCAACA........................................................................................................................................................... | 19 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................AGCAGC------AGGAACAGC-----------A---AC---------------------------A............................................................................................................................................................................................................................................................................................................................................................... | 19 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................AACAGCAGCA---GCAACAATATCAAC------------------AG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................AA------C----------AGCAGCAGCAACA------------------ATATCAACAG..................................................................................................................................................................................................................... | 26 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................................................................................A------C----------AGCAGCAGCAACA------------------ATATCAAC....................................................................................................................................................................................................................... | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................ACAGCAGCA---GCAACAATATCAAC------------------............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................AG----CAACA----------------------ACAGCAGCAGG..................................................................................................................................................................... | 18 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................CAGCA---GCAACAATATCAAC------------------AGCAA....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................ATCAACAGCAACAG----CAACA----------------------ACAGCAGCAG...................................................................................................................................................................... | 29 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................CAGCAGCAACA------------------ATATCAACAGCAA.................................................................................................................................................................................................................. | 24 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................AGCAGCAACA------------------ATATCAACAG..................................................................................................................................................................................................................... | 20 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................TATCAACAGCAACAG----CAACA----------------------ACAGCAG......................................................................................................................................................................... | 27 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................AGCA---GCAACAATATCAAC------------------AG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................AACAGC-----------A---AC---------------------------AA-CAGCAGCAGCA.................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................AA-CAGCAGCAGCAACA---------------------------------------------------------ACAAC................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................ACAGCAACAA------------------------------------------------------------------------------------CAGCAGCA-............................................................. | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACA------------------ATATCAACAGCAACAG----CAACA----------------------A............................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CAGCAACA---------------------------------------------------------ACAACAA------C----------AGCAGCA....................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CAACA---------------------------------------------------------ACAACAA------C----------AGCAGCAGCAA................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................CAACA------------------ATATCAACAGCAACAG----CAA........................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................CAACAG----CAACA----------------------ACAGCAGCAG...................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................ATCAACAGCAACAG----CAACA----------------------ACA............................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................AGCAACAG----CAACA----------------------ACAGCAGCA....................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................AACAGCAACAA------------------------------------------------------------------------------------CAGCAGCA-GCA---....................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................GCAGCAACA---------------------------------------------------------ACAACAA------C----------AGCAGC........................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GCAACA---------------------------------------------------------ACAACAA------C----------AGCAGCAGC..................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................A---------------------------------------------------------ACAACAA------C----------AGCAGCAGCA.................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................................................................................................................A------------------ATATCAACAGCAACAG----CAACA----------------------T............................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GCAGCAGCAACA---------------------------------------------------------ACAACAA------C----------AGC........................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................AGCAACA---------------------------------------------------------ACAACAA------C----------AGCAGCAGCAAC.................................................................................................................................................................................................................................................. | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................AA-CAGCAGCAGCAACA---------------------------------------------------------AC.................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................TATCAACAGCAACAG----CAACA----------------------ACAGC........................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACA------------------ATATCAACAGCAACAG----CAA........................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................ATATCAACAGCAACAG----CAACA----------------------AC.............................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................C---------------------------AA-CAGCAGCAGCAACA---------------------------------------------------------ACAACAA------C----------.............................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................ATATCAACAGCAACAG----CAACA----------------------ACAGC........................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................TATCAACAGCAACAG----CAACA----------------------ACA............................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................ACA------------------ATATCAACAGCAACAG----CAACA----------------------................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CA---------------------------------------------------------ACAACAA------C----------AGCAGCAGCAACA------------------A.............................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACA------------------ATATCAACAGCAACAG----CAAC....................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |