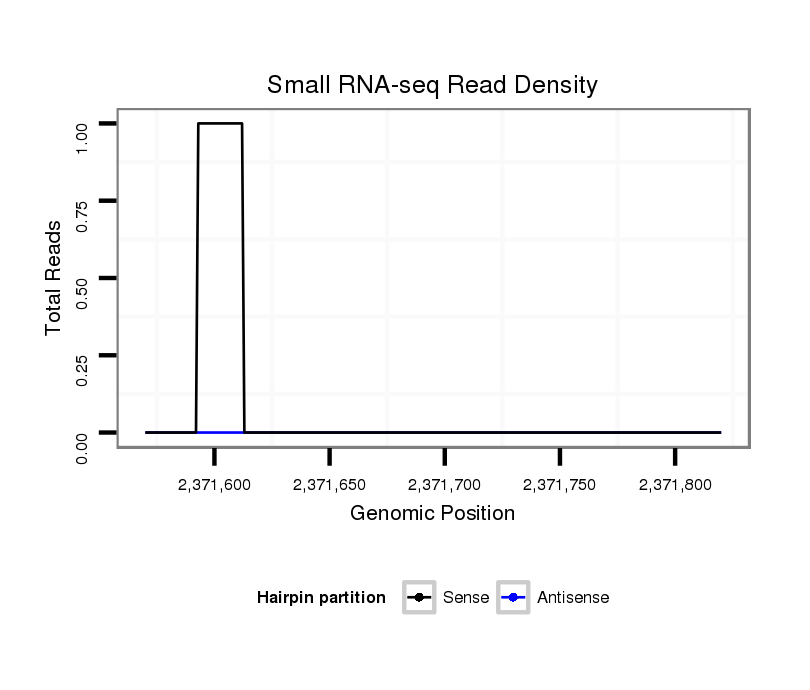
ID:dvi_2019 |
Coordinate:scaffold_12726:2371620-2371770 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ15379-cds]; exon [dvir_GLEANR_15692:2]; intron [Dvir\GJ15379-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATTGAAGAATGCCAAAATTCTAAACGCGGCCAATGGCCAATTCGAAAACGTAAGCTGTTTTACTGCATAACATTGTTCCGTCTAGATAATATTTACTTTTGCTGGAAATATTCTTCTTGAAATTCTTTCATATCTGATAAAAGCTTCATTTAATTAATCAGTTTTTATTACATAGGTTAGCGGGCGATAATTTAGTACTCTAGTTAAAATTTATTAGTAGCAACTATTTAACATGCCTTTTACTAGGGCA **************************************************.((((.((((...((((.((((.........(((((..((........))..)))))...........(((((...)))))(((.(((((((((((..(((.....))).))))))))))).))).))))))))..)))).))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
M061 embryo |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|
| .......................AACGCGGCCAATGGCCAATT................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................AACCATTAACCATGCCTTTT......... | 20 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| ...................................................TGAGCTCTTTTACTGCATAT.................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
|
GTAACTTCTTACGGTTTTAAGATTTGCGCCGGTTACCGGTTAAGCTTTTGCATTCGACAAAATGACGTATTGTAACAAGGCAGATCTATTATAAATGAAAACGACCTTTATAAGAAGAACTTTAAGAAAGTATAGACTATTTTCGAAGTAAATTAATTAGTCAAAAATAATGTATCCAATCGCCCGCTATTAAATCATGAGATCAATTTTAAATAATCATCGTTGATAAATTGTACGGAAAATGATCCCGT
**************************************************.((((.((((...((((.((((.........(((((..((........))..)))))...........(((((...)))))(((.(((((((((((..(((.....))).))))))))))).))).))))))))..)))).))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
V047 embryo |
SRR060665 9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060656 9x160_ovaries_total |
SRR060673 9_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................GTATTGTCACGAGGCAGAT...................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................CTATATAAGAAGAAATTTAAGAT........................................................................................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AGTAAATCAATTAGTCAAA...................................................................................... | 19 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| GTAGCTTCTGACGGTTTTA........................................................................................................................................................................................................................................ | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................TGAAAACGACCTTTAACA.......................................................................................................................................... | 18 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| ......................................................CGGCAAAATGAGGTAGTGTA................................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TAAGAAGAACTTTAGGAG........................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:2371570-2371820 + | dvi_2019 | CATTGAAGAATGCCAAAATTCTAAACGCGGCCAATGGCCAATTCGAAAACGTAAGCTGTTTTACTGCATAACATTGTTCCGTCTAGATAATATTTACTTTTGCTGGAAATATTCTTCTTGAAATTCTTTCATATCTGATAAAAGCTTCATTTAATTAATCAGTTTTTATTACATAGGTTAGCGGGCGATAATTTAGTACTCTAGTTAAAATTTATTAGTAGCAACTATTTAACATGCCTTTTACTAGGGCA |
| droMoj3 | scaffold_6654:475293-475356 + | CCGTTAACTATAGTGAGGTCCTCATAGCGGCAACAGCCCGTTTTGGTAATGTAAGTGCTTTGCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15110:7721853-7721909 - | CATTTGACTTTGGCAATATTCTCATTACAGCCACGGCTCATTTTGGCAATGTAAGTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13337:2012378-2012434 + | CCTTCGCCTACAGAGACATAGTTATAGCCGCCACAGTTCAGTTCCGTAATGTAAGTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000454113:640000-640048 + | CATAC------AAGGATATACTGGAAGCTGGCACCTACCAGTTTCGCAATGTAAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409007:256858-256861 - | CATT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/17/2015 at 04:54 AM