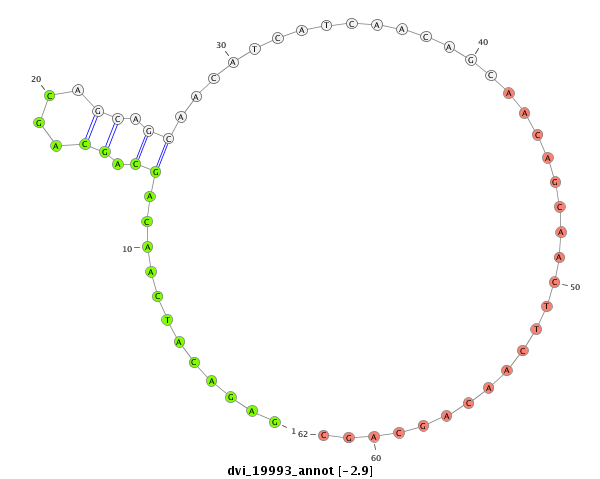
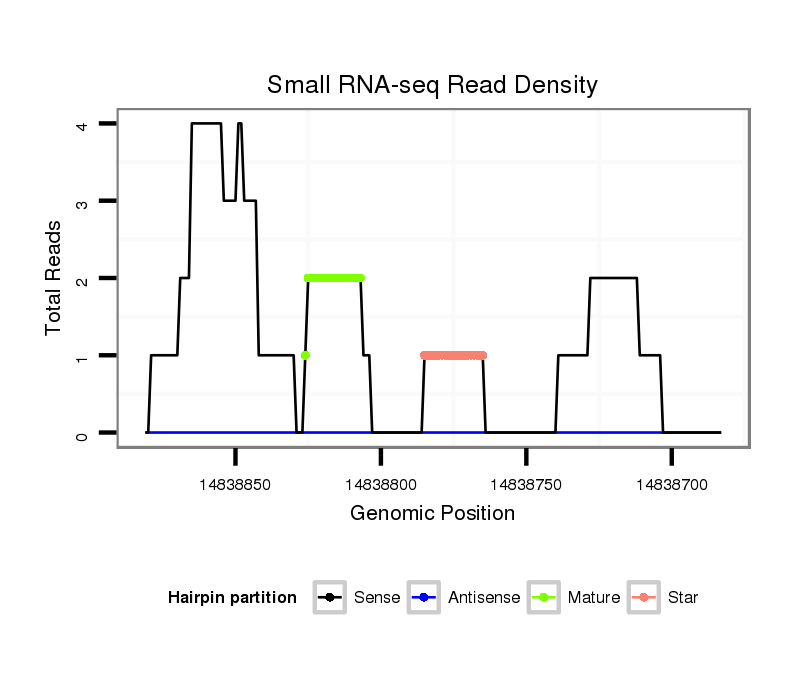
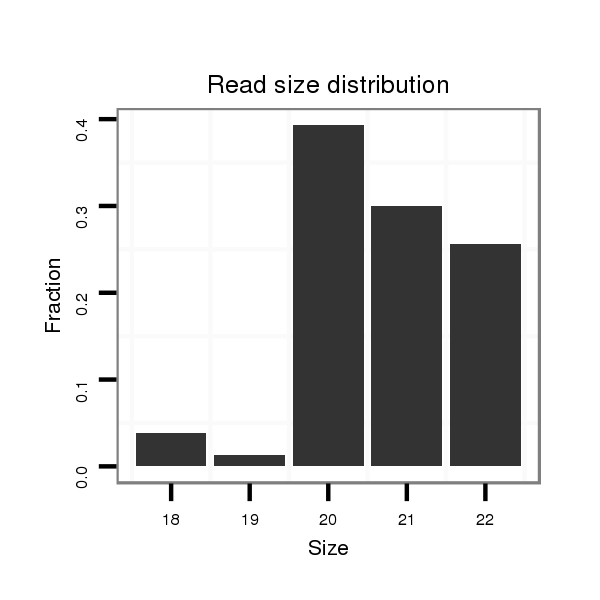
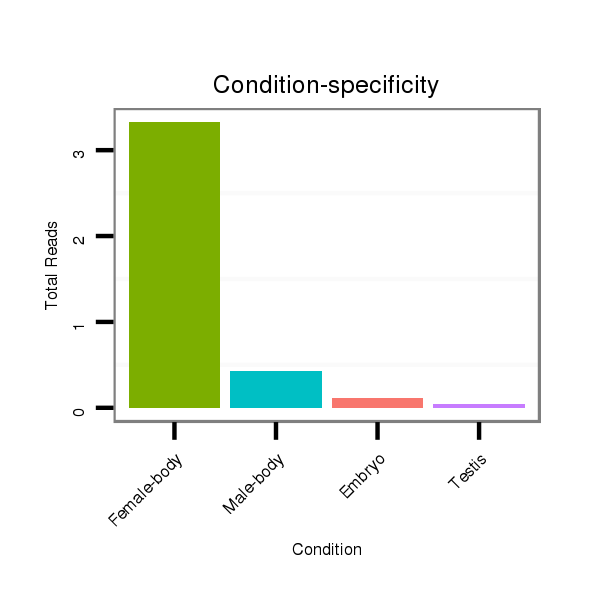
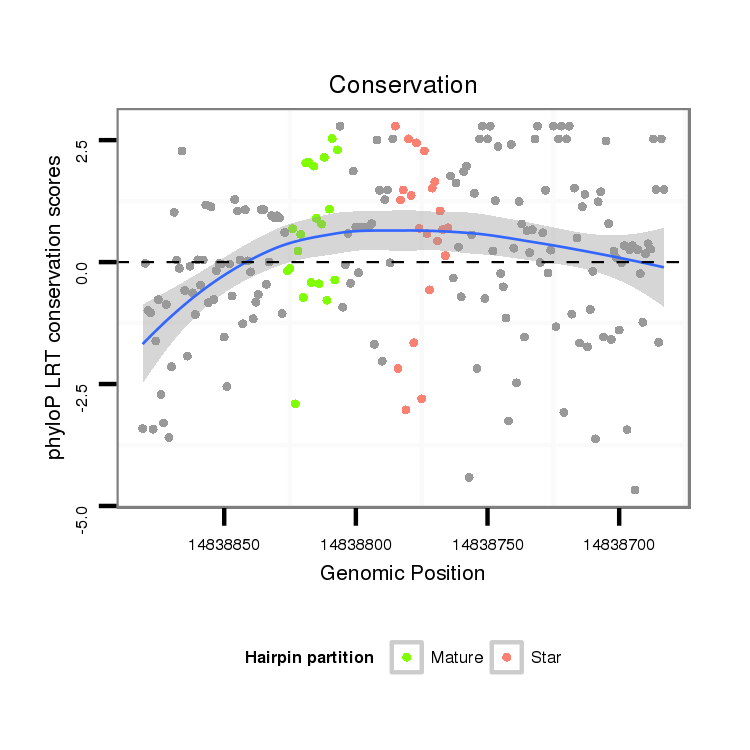
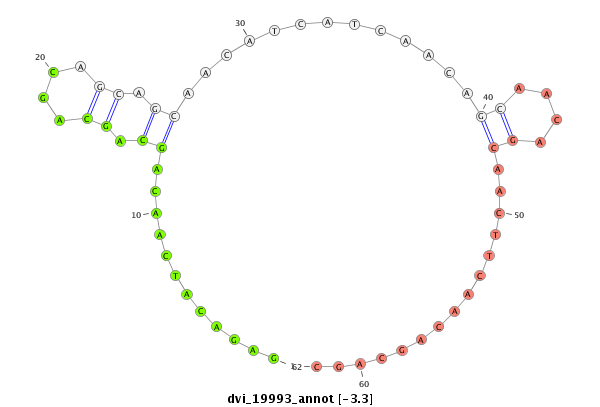
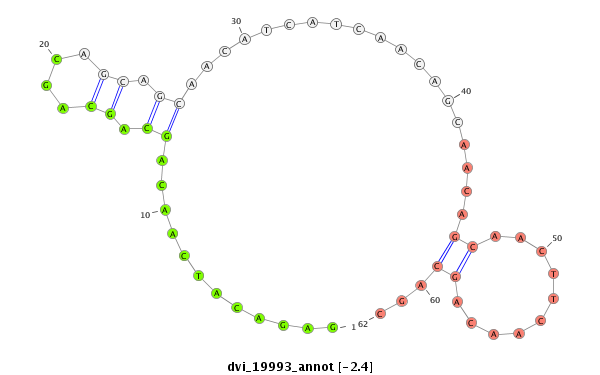
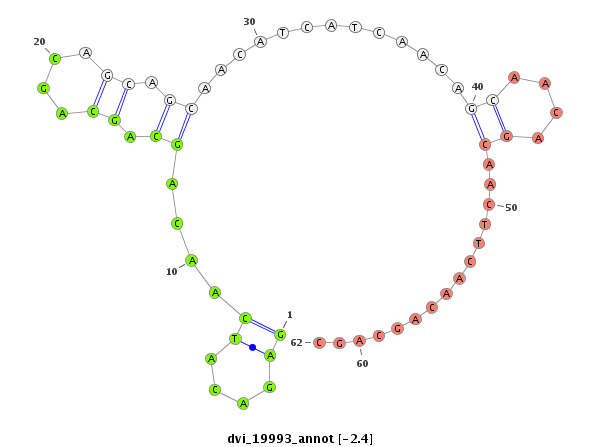
ID:dvi_19993 |
Coordinate:scaffold_13049:14838733-14838831 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -3.3 | -2.4 | -2.4 |
 |
 |
 |
exon [dvir_GLEANR_11786:6]; CDS [Dvir\GJ11771-cds]; CDS [Dvir\GJ11771-cds]; exon [dvir_GLEANR_11786:5]; intron [Dvir\GJ11771-in]
| Name | Class | Family | Strand |
| TART_DV | LINE | Jockey | - |
| ##################################################---------------------------------------------------------------------------------------------------################################################## GCACTTCATCTTCGGCTACCCTGAAGCGGACTCCCTCGGCTAGTACCGTTGTGGAGAGACATCAACAGCAGCAGCAGCAGCAACATCATCAACAGCAACAGCAACTTCAACAGCAGCAGCAACATCAACAGCAGCAACTTCAGCAGCAGCAAGAACAGCAACAGCGACGCACTCAACAAAAACATCAGTCGGAGCAACA *******************************************************............((.((....)).))....................................********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M027 male body |
SRR060685 9xArg_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060681 Argx9_testes_total |
SRR060666 160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060657 140_testes_total |
V047 embryo |
SRR060682 9x140_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................TACCCTGAAGCGGACTCCCTCGG................................................................................................................................................................ | 23 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................GAGACATCAACAGCAGCAGC............................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GCAGCAGCAAGAACAGCAACAGCGACGC............................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................AACAGCAACTTCAACAGCAGC.................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................AGACATCAACAGCAGCAGCAGC......................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AACAGCAACAGCGACGCACTCAACA..................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CGGCTACCCTGAAGCGGACTCC..................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CCCTCGGCTAGTACCGTTGT................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GACCCTGAAGTGGACTCC..................................................................................................................................................................... | 18 | 2 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..ACTTCATCTTCGGCTACCCTGAAGC............................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............CGACCCTGAAGTGGACTCCA.................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TCAACAGCAGCAGCAACATC......................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CAGCAGCAGCAAGAACAGCAACA.................................... | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................ACAGCAGCAGCAGCAGCA..................................................................................................................... | 18 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATCAACAGCAGCAGCAGCAGCAACATT................................................................................................................ | 28 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................AGTACAGTTATGGAAAGACA.......................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................GCAACATCAACAGCAGCAAC............................................................. | 20 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CATCAGCAACTTCAACCGC..................................................................................... | 19 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CAGCAACGACAACAGCAGCAACTTC.......................................................... | 25 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................AGCAGCAACATCAACAGCAGC................................................................ | 21 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................AGCAGCAACATCAACAGCAG................................................................. | 20 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GCAGCAACATCAACAGCAGCA............................................................... | 21 | 0 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................AACAACTTCAACAGCAGCAGCA.............................................................................. | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................CATCAACAGCAGCAGCAGC......................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................ACAGCAGCAGCAGCAGCAACA.................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................AACACAAGCAACTTCAGCAGCAG.................................................. | 23 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................GCAGCAACATCAACAGCAGC................................................................ | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGTGAAGTAGAAGCCGATGGGACTTCGCCTGAGGGAGCCGATCATGGCAACACCTCTCTGTAGTTGTCGTCGTCGTCGTCGTTGTAGTAGTTGTCGTTGTCGTTGAAGTTGTCGTCGTCGTTGTAGTTGTCGTCGTTGAAGTCGTCGTCGTTCTTGTCGTTGTCGCTGCGTGAGTTGTTTTTGTAGTCAGCCTCGTTGT
**********************************************************************************............((.((....)).))....................................******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M027 male body |
SRR060683 160_testes_total |
SRR060667 160_females_carcasses_total |
M028 head |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060677 Argx9_ovaries_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060670 9_testes_total |
SRR060671 9x160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TTGTCGTCGTTGAAGTCT....................................................... | 18 | 1 | 6 | 0.33 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................CGTCTTAGGGAGCCGATCAC.......................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TCGAAGCCGATAGGACTCC............................................................................................................................................................................. | 19 | 3 | 11 | 0.27 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......TCGAAGACGATGGGACTCC............................................................................................................................................................................. | 19 | 3 | 12 | 0.25 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....AAGGAGAAGCCGCTGGGA................................................................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....AGGAGAAGCCGCTGGGAC................................................................................................................................................................................ | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TCGTCGTCGTTGTAGTAGTTA.......................................................................................................... | 21 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGTAGTTGTCGTCTCTGAA........................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCGTGGCCGCTGCGTGAG......................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GAAGGAGAAGCCGCTGGGAG................................................................................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................GCGCTGTAGTAGTTGCCGTTGT................................................................................................... | 22 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TGTTCTTGTAGTCAGACT...... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................GCCTTAGTGAGCCGATCAC.......................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......TAGAAGACGATCGGACTCC............................................................................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................TTGTAGTAGTTGACCTTGT................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:14838683-14838881 - | dvi_19993 | GCA-------------------------------CTT----------------------C--A-TCTTCGGCTACCCTGAAG---CGGACTCCCTCGGCTAGTACCGTTGTG--------------------------------GAGA---------------------------------------GAC---A----------------TCAACAGCAGCAGC---------AGC------------------------AGCAACATCATCAACAGCAACAGCAACTTCAACAGCAGCAGC---AACA------------------T------CAACAGCAGCAACT------------TCAGCAGC---AGCAAGAACAGCAA---CAGCGACGCAC----TCAACAAAAA------------CA------------TCAGTCGGAGCAACA |
| droMoj3 | scaffold_6680:19620127-19620316 - | GCG-------------------------------CTT----------------------C--G-TCTTCGGCTACATTAAAA---CGAACTGCCTCGACCATAACCGTTGTG--------------------------------GAGA---------------------------------------------A-------------ACAGCAACAGCAGCAACTGC---AGCAGA------TG---------------------------CAACAGCAACAGCAACAGCTG---C---------AACA------------------A------CAACAGCAGCAACA---------ACAACTACAACA---GCAGCAGCAACAACAACA---TGAACA----ACAGCGACGA------------AC------------ACAACTCAAGCAACA | |
| droGri2 | scaffold_14612:1371-1552 + | CAG-------------------------------CAG----------------------C--A-GCGGCAGCAGC---------------------------------------AGCAGC---GGCAGCAG-------------------------------------CAAAA---------ACGGCAG---CA----------------------CCAACAGC---AGCAGCAGC------------------AGCAGCGTC------AACATCAGCAGCAGCAGCTGCAGCAGC---------------AGCAGCAGTT------A------AAACAGCAGC---A------------GCATCAGCA---GCAGCAAAAACAGCAGCA---GCAACA----GCAGCAGCAG------------CA------------TCAGCAGCAGCAGCA | |
| droWil2 | scf2_1100000004888:1009828-1010013 - | ACA----------------------ACAAC------------------------------------AGCAGCAAC-------A-----------------------------ACAGCAGCAGCAGCAACAG-------------------------------------CAACA---------ACAGCAG---CA----------------------GCAACAAC---AACAGCAGC------------------AACAACAAC------AGCAGCAGCAGCAACAGCAACAACAGC---------------AGCAGCAACA------A------CAACAGCAGC---A------------ACAACAACA---GCAGCAGCAACAACAGCA---ACAGCA----ACAACAACAG------------CA------------GCAGCAACAGCAACA | |
| dp5 | Unknown_singleton_1849:5259-5444 + | GCA----------------------GCAAC--------------------------------A-TCTGCAGCA---------------------------------------GCAACAGCAGCAACAAC-------------------------------------AGAAGC---------------------A-------------GCAGCAACAGCAGCAGC---AGCAGCAGG------------------TGCAGCAGC------AACATCAGCAACAGCAACATCAGCAGC---------------AACAGCAGCA------A------AAGCAACATC---A------------GCAGCAACA---GCAGCAGCAGCAGCAACA---ACAGCA----ACAACAGCAA------------CA------------TCAGCAACAACAACA | |
| droPer2 | scaffold_4:6701389-6701560 + | CGG----------------------AGACC------------------------------------------------------------------------------------------------------------------ATGAAG---------CGACCCATCC------------------ACC---A-G------------CAGCAACAACATCAGCTGC---AGCAGCAGCAACAA------------------C------AGCAGCAACATCAACTGCAGCAG---CAGCATC---AGCA------------------G------CAGCAACAGCAGCT------------GCAGCAGA---AGCAGCAACAGCAA---CAACAGCAGCA----GCAACATAAA------------CA------------TCAACAGCAACAACA | |
| droAna3 | scaffold_13337:9798605-9798798 - | GCAATCGATGAGAGCCTGCTCGAGACTTTCTCAA---CGCCG----------CTGCTCAGTCCTTTG----------------------------------------------------------------GAGATAA-AGACCGAAAAGCA---GCAG---------------------------------CGA-------------CAGCAGC---------------AGCAGC------AA---------------------------CAACATCAACACCAACAC------CAGCAGC---AACA------------------G------CAACAGCAGCAATA------------C---CAGCAGCAGCAGCAACAGCAA---CAG------CA----CTATCAGCAA------------CA------------TTATCAACAGCAGCA | |
| droBip1 | scf7180000396544:101354-101539 + | GCA----------------------GCATC--------------------------------A-GCAACTACT---------------------------------------GTCGCAGCA---GCAGCTC----------------------------------TTGT---C---------------G-------------CAG---CAGCAACAACAGCAGC---AGCAGCAATACCAGCAG------------------C------CGCAACAGCATCAGCAGCATCAC---T---------------TC------------------------CAGCAACAACAGCAGCAGCAGCCGCTTTTGCA---GCAGCAACAACACGCACA---GCAGCA----GCAGCAGCAA------------CA------------TCTGCCACAGCAGCA | |
| droKik1 | scf7180000302685:196422-196619 + | ACA----------------------TCACC------------------------------------AGCAGCAAC-------AA--CAACAGC---------------------ATCTAC------AGCAG-------------------------------------CAACA----------------------------------------GCA---GCAGCTGC---AGCAAC------ATCTACAGCAGCAACAGCAGCAGCATCATCTACAGCAGCAGCAGCATCTACAGCAAC------------AGCAGCA---------T------CTACAGCAGCAGCA------------GCAGCAGCA---GCATCTACAGCAGCAGCA---GCAGCA----GCAGCAGCAT------------CA------------TCTCCAGCAGCAGCA | |
| droFic1 | scf7180000454096:1059357-1059565 + | GCA----------------------GTAGT----AGT--------CATGTTGG------T--G-CTGGTAGCGGTAGTCAAG---CAAATAGTGTC---------------------------------------------------------------------A------AGCATCTGCA---GAATA---A----GCAAAAGCAACAACAACT---ACAAC---AACAGCAAA------AG---------------------------CAACAGCAACTGCAACAG------CAAAAGC---AACA------------------A------CAACAACAGCAACC------------GCAACAGC---TACAGCAACAGCAA---CAGCATCACGATGTTGCAGCAGCAA------------CA------------TTAGCAGCAGCAACA | |
| droEle1 | scf7180000491214:2315505-2315699 - | GCA-------------------------------TAT---CGC------------CGCAGGCCCTTG---GCAGC-------------------------------------------------------------AATTCGCCCAAGAGCAATTGCAG---------------------------------CAA-------------CAGCAGCAGCAGCAGCAGT---AGTAGC------AG---------------------------CAGCAGCAGCATC---------------------AGCA------------------G------CAGCAGCAGCAACA------------G---CAGCAGCAGCAGCAACAGCAG---CATCAGCAGCA----GCAACAGCAACGCCAGCGATCAGCCCACTCACAGCAACATTCCGCGCAGCA | |
| droRho1 | scf7180000779872:456932-457121 - | TCA----------------------AGGCC----------------------------------TATACACCCAGC-------A--CAACGCCTTCTCCTCCAGCCACTGTG-------------------GCGGGAA-AACCCAAGCAACA---GCAA---------------------------------CAG-------------CAAC------------------AGCAAC------AG---------------------------CAACAGCAGCAGCAGCAGCAG---CAGCAAC---AGCA------------------G------CAGCAGCAGCA---------------ACAGCAGCA---GCAGCAACAACAGCAGCA---ACAACA----GCAGCAACAA------------CA------------GCAGCAGCAGCATCA | |
| droBia1 | scf7180000298389:213008-213188 - | GCT-------------------------------CCACTTCAGTGCAGGTCGGTGCTCCGCCGCCCA-------------------------------------------------------------------------------CA---------------------------------------AACAACAA-------CAACAACAACAGCA---GCAGCAGC---AGCAGC------AA---------------------------CAGCAGCAACAACAACAGCAG------CAGC---AACA------------------G------CAGCAGCAGCAACA------------ACAGCAAC---AGCAGCAGCAGCAA---CAACAACACAC----CCAACGGTGC---------G--CA------------GCAGCAGCAGCAACA | |
| droTak1 | scf7180000415785:203976-204159 - | AGA----------------------CTTTCTCGA---CCCCA----------TTGCTCAGTCCCCTG----------------------------------------------------------------GAGATAA-AGACGGAGAAA----------------------------------------------------------CAGCAGCAGCAGCAGCAGCAACAGCAGC------GA---------------------------CAACAGCAGCAGCAACATCAG------CAGCAACAGC---A---------------A------CACCAGCAGCA---------------ACATCAGCA---GCAGCAATACCAGCAGCA---ACACTA----TCAGCAACAT------------TA------------TCAGCAGCAGCAGCA | |
| droEug1 | scf7180000409246:298182-298373 + | TCA----------------------TCATC--------------------------------A-TCATCATCA---------------------------------------CCATCAACA---TCATCAC-------------------------------------AGTCAGCATCTACAACTCCAA---CAA-------CAACAACAAC------------------AACAGC------------------AGCAACAAC------AACAACGGCAACACCAACAACAG---CCGC------------TACACCA---------A------CAACAACAGCAATT------------GCAACACC---AGCAGCAACAACAA---CAACAACACCA----CCAACAACAA------------CA------------ACACTACCAACAACA | |
| dm3 | chr3L:7824164-7824345 - | GCC-------------------------------TTG----------------------C--A-GCAGCAGCAAC-------AAACCGCCGCC------------------------------------------------------------------------CTGC------------------ACC---A-TCAGCAACAG---CAGCA---GCATCAACAGCAGCAGCAAC------------------------------------ACCAACAGCACC---------------------TACAGCAACACCA---------TCAAGCGCACCATCAGCATGTCCT---------GCAGCAGC---AGCAGCAGCAGCAA---CAGCAGCACTC----CCAGCATCAG------------CA------------TCAGCAGCAACATCA | |
| droSim2 | x:10052417-10052623 + | TGT-------------------------------CGC----------------------C--A-TCTCCGCCGCTCAACGAT---CAGATCAGCT-TGCTATTCCCC-------------------------------------AAGTGC---------CGACC---------------------CAAACAGCAG-------CAGCAGCAGCAGCA---GCAGCAGC---AGCAGC------AG---------------------------CAGCAGCAGCAACAGCAG------CAGCAGC---AACA------------------G------CAACAGCAGCAACT------------TCAACCAG---CACAACAGCAGCAG---CAGCAGCATGTCCTTCCCACA--GA------------CG---------TGAATTCCAGCTGCAACA | |
| droSec2 | scaffold_0:130592-130770 - | GCC-------------------------------TTG----------------------C--A-GCAGCAGCAAC-------AAACCGCCGCC------------------------------------------------------------------------CTGC------------------ACC---A-T------------CAGCAACAGCAGCAGCAGC---AGCATC------AA---------------------------CAGCAGCAACACCAACAGCAC---C---------TACAGCAACACCA---------TCAAGCGCACCATCAGCATGT------------CCTGCAGC---AGCAGCAGCAGCAA---CAGCAGCACTC----CC---AGCAT------------CA------------GCATCAGCAGCAACA | |
| droYak3 | 3L:4195061-4195253 + | TCG-------------------AGACATTCTCGA---CCCCA----------TTGCTCAGTCCCCTG----------------------------------------------------------------GAGATAA-AGACCGAGAAGCA---GCAG---------------------------------CGA----------CAACAGCAGCA---CCACCACC---AACAC---------------------------C------AGCACCAGCAGCAGCAACAACAG---CAGCAAC---AGCA------------------G------CAACACCAGC---A------------GCAACATCA---GCAGCAATACCAGCAGCA---ACACTA----TCAGCAACAT------------TA------------TCAGCAGCAGCAGCA | |
| droEre2 | scaffold_4929:4974122-4974309 + | GCA-------------------------------CAG----------------------C--A-GCAACAGCAGC---------------------------------------AGCAAC---AACATCGC-------------------------------------TAATAACAGCTATA----------------------------------AGAGAAGC---AGCAGCAGC------------------GGCAACAAC------AGCACTAACAGAAGCAGCAGCAGCAAC---------------AACAGCAGTTTAAACAT------CAGCAACAAGAATG------------ACAGCAAC---AGCACTAACAGCAA---CAACAACAGCC----GC---AGCGA------------CA------------ACATCGACGGCGACG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 09:51 PM