
ID:dvi_19979 |
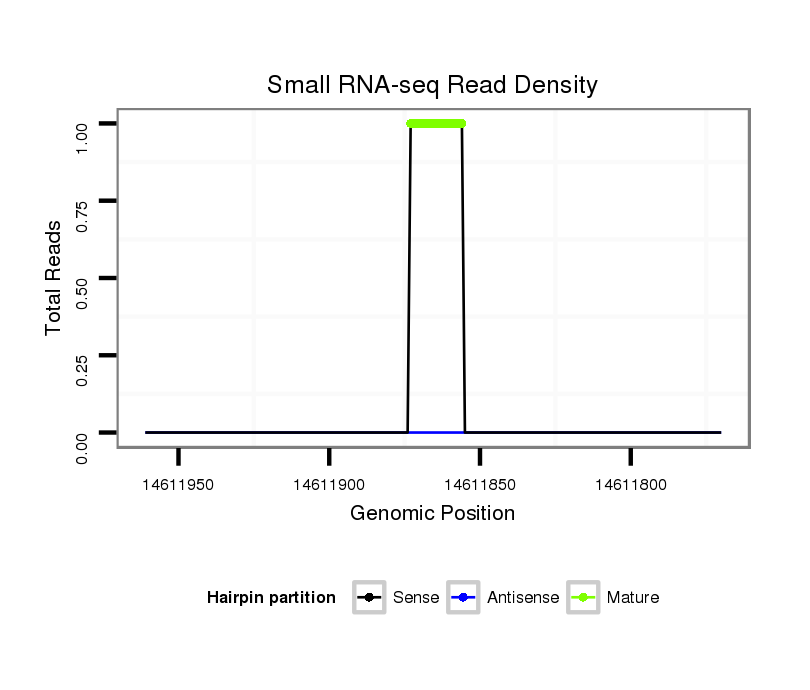
Coordinate:scaffold_13049:14611820-14611911 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.4 | -9.3 | -9.3 |
 |
 |
 |
CDS [Dvir\GJ11776-cds]; CDS [Dvir\GJ11776-cds]; exon [dvir_GLEANR_11791:2]; exon [dvir_GLEANR_11791:1]; intron [Dvir\GJ11776-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------################################################## AAGCTGAGAATATGAGAACCCACGGCAAACTGACCGTGGACTTGAGTATTGTAAGTCGATAAATAGAGCATACAAAGTACTTTCGGATCCAGATCTGCGCAGAGACTATGATCGAATCTATAATGCCCATAATAATCAAAAGAAAAGCCAAGGAGGCCAGTCACAGGGCCATCAACAGGAGAATGCGAAATC **************************************************.................((((.....(((..((((.(((.((.(((......)))))..)))))))..))).))))................************************************************** |
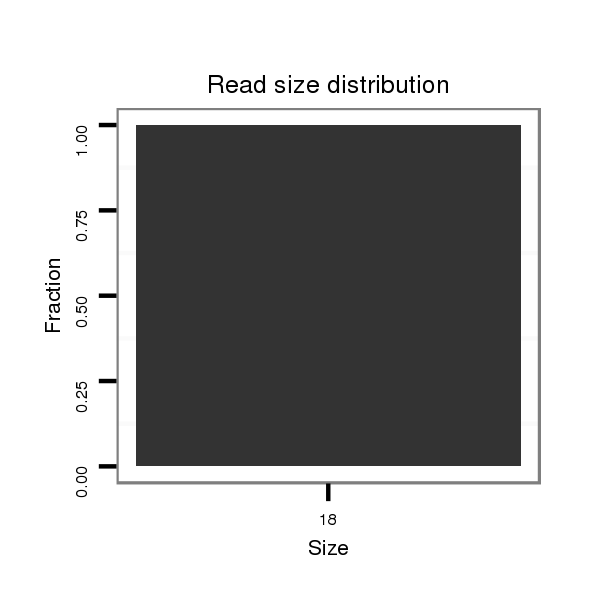
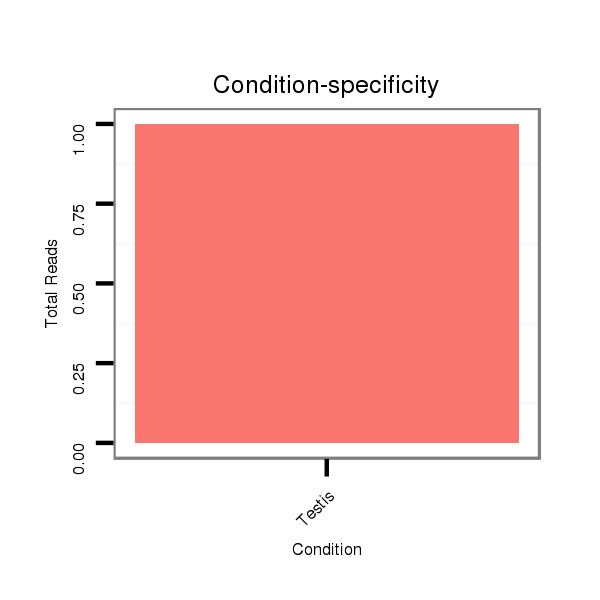
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
M028 head |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................CCAGATCTGCGCAGAGAC...................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CGTGGACTTAAGCATTATA........................................................................................................................................... | 19 | 3 | 20 | 0.75 | 15 | 0 | 7 | 0 | 3 | 2 | 2 | 1 |
| .......................GGCAGACAGACCGTGGAC....................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TTCGACTCTTATACTCTTGGGTGCCGTTTGACTGGCACCTGAACTCATAACATTCAGCTATTTATCTCGTATGTTTCATGAAAGCCTAGGTCTAGACGCGTCTCTGATACTAGCTTAGATATTACGGGTATTATTAGTTTTCTTTTCGGTTCCTCCGGTCAGTGTCCCGGTAGTTGTCCTCTTACGCTTTAG
**************************************************.................((((.....(((..((((.(((.((.(((......)))))..)))))))..))).))))................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
V053 head |
|---|---|---|---|---|---|---|---|
| ..............TCTTGGCTGCCATTTGTCTG.............................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 |
| ........................CGATTGAATAGCACCTGAAC.................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 |
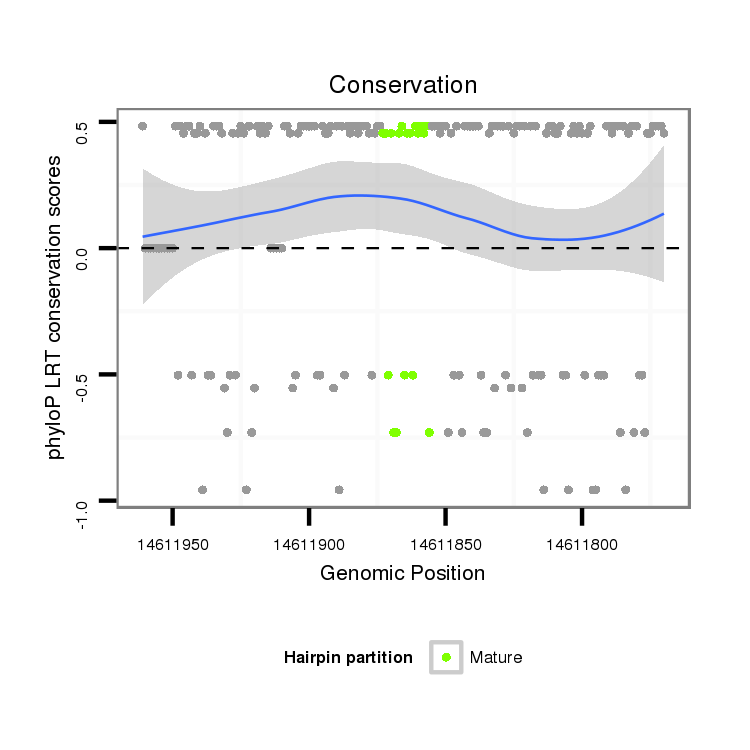
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:14611770-14611961 - | dvi_19979 | AAGCTGAGAATATGAGAACCCACGG--CAAACTGACCGTGGACTTGAGTATTGTAAGTCGATAAATAGAGCATACAAAGTACTTTCGGATCCAGATCTGCGCAGAGACTATGATCGAATCTAT---AATGCCCATAATAATCAAAAGAAAAGCCAAGGAGGCCAGTCACAGGG---------------------------------------------------------------------------------------CCATCAACAGGAGAATGCGAAATC |
| droMoj3 | scaffold_6680:19359436-19359703 - | A-----------TAAGAATCCAGGAAATAAACATGCTGTGCAAATGAGT-----AAGATGATAAATGAAGCAAAGAGAGTACTTTCAGATCCGGCGCTACGTAGAGAATATGATAGGACATATATTAATAAACAAAATGAACAATATAGAGAGCAAGGAAAGCAGTCGCACCACTCGCAGAAGAACACAAAATCGAACTATAATCATGATCAAAATAATAGACAGGGCACGACAAAAGACGACAAGTCAAATGGACGCCATTATCAAAACGATAGCTCGAAATC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/17/2015 at 05:08 AM