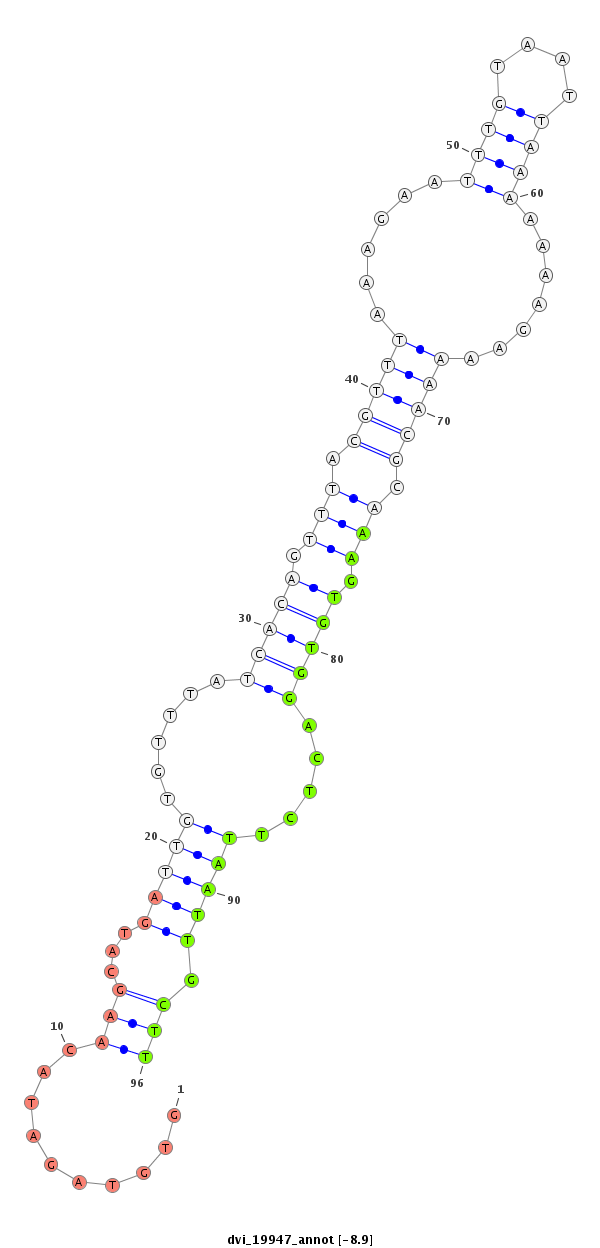
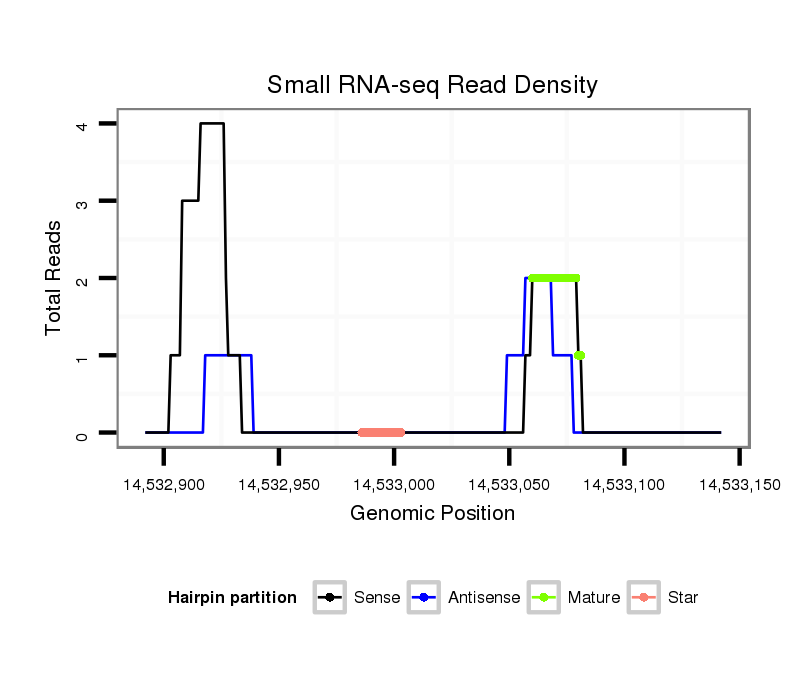
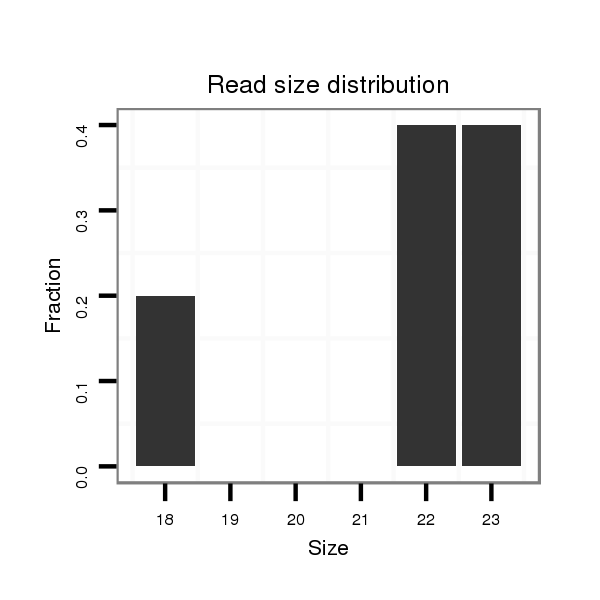
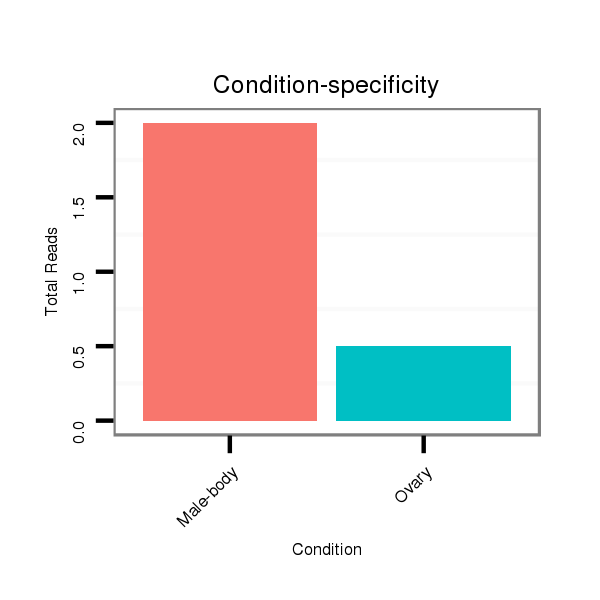
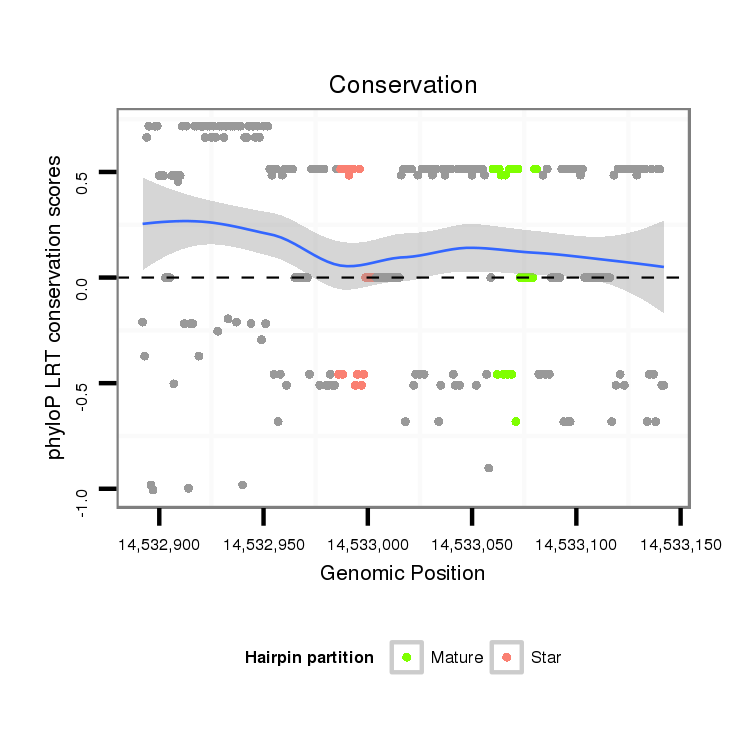
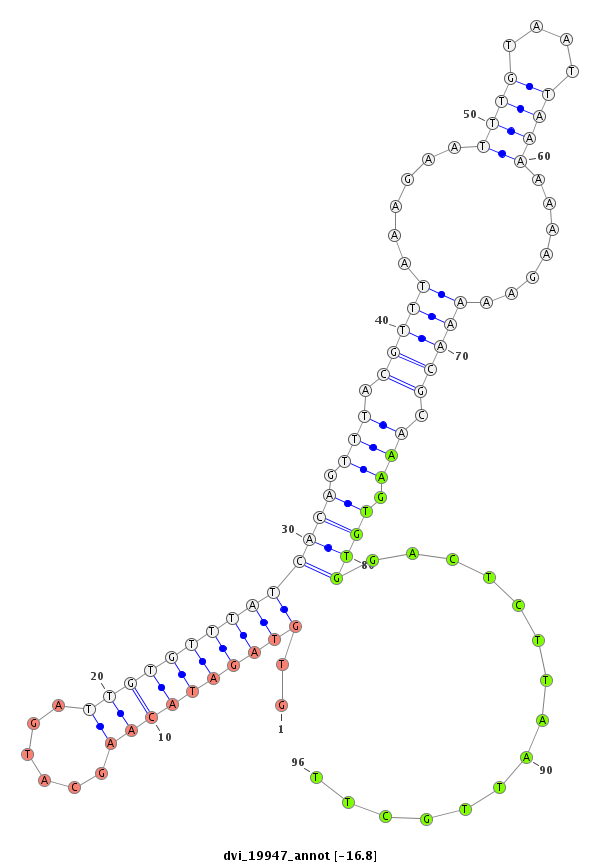
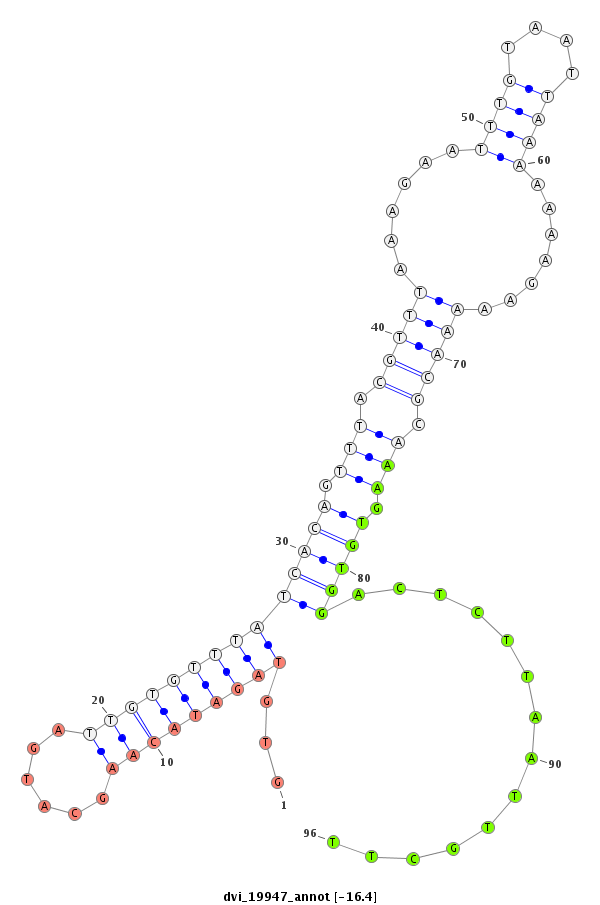
ID:dvi_19947 |
Coordinate:scaffold_13049:14532942-14533092 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.8 | -16.6 | -16.6 | -16.4 |
 |
 |
 |
 |
exon [dvir_GLEANR_13448:1]; CDS [Dvir\GJ13565-cds]; intron [Dvir\GJ13565-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| mature | star |
| ----------------------------######################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGCTGTAAATTATTACAGAACAGACAACATGATGTCAAAGAGATTATTGGGTGAGTGCAAATGTTGCCTTATAATATATATAAATAATAACATTGTGTAGATACAAGCATGATTGTGTTTATCACAGTTTACGTTTAAAGAATTTGTAATTAAAAAAAGAAAAACGCAAAGTGTGGACTCTTAATTGCTTTACCACCCGCAAGAGGAAAACAACAATGTTTGCGACGAATAAATTTTGATTATCTTAAATT **********************************************************************************************..........(((...(((((......(((((.(((.(((((......((((....)))).......))))).))).))))).....))))).)))************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M047 female body |
V116 male body |
M027 male body |
SRR060659 Argentina_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060683 160_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......AACTTATTACAGAACAGACAACATGAT.......................................................................................................................................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGAACAGACAACATGATGTC....................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................GTCAAAGAGATTATTGGCTCAGA................................................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AAGTGTGGACTCTTAATTGCTT............................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTGAGTGGTAATGTTGCC....................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGAACAGACAACATGATGT........................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AGGGATTATTGGGTGAGTGCA................................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................AGGAGATTGTTGGGTGAGTGC................................................................................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TTGTTACAGAACAGACAACA.............................................................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........ATTACAGAACAGACAACATGATGT........................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTGTGTGCAAAGGTTGCC....................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................GCAAAGTGTGGACTCTTAATTGC............................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................CAACATGATGTCAAAGAG................................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................CGGGTGAGTGGTAATGTTGCC....................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGAGAGTGTTAATGTTGCC....................................................................................................................................................................................... | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................ACATGATGTCATAGAGAGAATT........................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GTGTAGATACAAGCATGA........................................................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TGGGAGAGTGGTAATGTTGCC....................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTGAGTGGTTATGTTGCC....................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CGGGTGAGTGTTAATGTTGCC....................................................................................................................................................................................... | 21 | 3 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TAAAGAATTTGTAACAAAAAAAAAAAA......................................................................................... | 27 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................TGGGTGAGTGGTAAGGTTGC........................................................................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GGGTGAGCGTGAATGTTGCC....................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTGAGCGCCGATGTTGC........................................................................................................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTCAGTGGTAATGTTGC........................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GATACAGGCGTGATCGTGT..................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGAGAGTGTTAATGTTGC........................................................................................................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTGAGTGGTGATGTTGC........................................................................................................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGGGTGATTGGTAATGTTGC........................................................................................................................................................................................ | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................ATAGATACAAGGAGGATTG........................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TCGACATTTAATAATGTCTTGTCTGTTGTACTACAGTTTCTCTAATAACCCACTCACGTTTACAACGGAATATTATATATATTTATTATTGTAACACATCTATGTTCGTACTAACACAAATAGTGTCAAATGCAAATTTCTTAAACATTAATTTTTTTCTTTTTGCGTTTCACACCTGAGAATTAACGAAATGGTGGGCGTTCTCCTTTTGTTGTTACAAACGCTGCTTATTTAAAACTAATAGAATTTAA
*************************************************************..........(((...(((((......(((((.(((.(((((......((((....)))).......))))).))).))))).....))))).)))********************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060654 160x9_ovaries_total |
SRR060668 160x9_males_carcasses_total |
SRR060683 160_testes_total |
M047 female body |
SRR060679 140x9_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060680 9xArg_testes_total |
V047 embryo |
SRR060678 9x140_testes_total |
SRR060689 160x9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................TCTTTTTGCGTTTCACACCT.......................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TGTACTACAGTTTCTCTAATA............................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CGTTTCACACCTGAGAATTAA................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................ACTGAGAATTAACGAAAT........................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ATTTATTTTTTTCTTTTGGCGTCT................................................................................. | 24 | 3 | 5 | 0.60 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................GTAAGGAAATGGTGGGCGT.................................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........AGAAAGTCTTGACTGTTGTA............................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.30 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| ......................................................................................................................................................................................GTAAGGAAATGGTGGGCG................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CTAACACAAATAGTGACGAGT........................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TTTTTTTTTACAAACGCTGATTA..................... | 23 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ATTTATTTTTTTCTTTTGGCGT................................................................................... | 22 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................ATGATAATTAACGAAATGGTA....................................................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........TAAGAATGTTTAGTCTGTTG............................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:14532892-14533142 + | dvi_19947 | AGCTGTAAATTATTACAGAACAGA--CAACATGATGTCAAAGAGATTATTGGGTGAGTGCAAATGTTGCCTTATAATATATATAAATAATAACATTGTGTAGA--------------------TACAAGCATGATTGTGTTTATCACAGTT----TACGTTTAAAGAATT------------------------TGTA-------------------------ATTAAAAAAAGAAAAACGCAAAGTGTGGACTCTTAATTGCTTTACCACCCGCAAGAGGAAAACAACAATGTTTGCGACGAATAAATTTTGATTATCTTAAATT |
| droMoj3 | scaffold_6680:12766668-12766727 - | AACTAGAAATT---ATAGAATATATTTAATATGATGTCGAAGAGATTATTAGGTAAGTGCAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15110:15699845-15700107 - | GACTTAAA-----------ACAAT--CAATATGATGTCAAAGAAATTGTTTGGTGAGTGAATATGCTTTCTAATA-------CAAATTATTTTTATATATAGATTTCTATAATTTTTTTTATATTTATA-----------------CATTTGTTGTTTGCTCAAAGAAGAAGAAAATGAAACAGCATTTTTATATGTAAATATATATTTTGAAGGAAAACAAGACAATAAAAAGATAAACAG-AAATGCGAATTAT-------TTCGCTAT-----ATATTAAAACA-------------AGTACATATTTTGATTAGTTCCAAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 05:10 AM