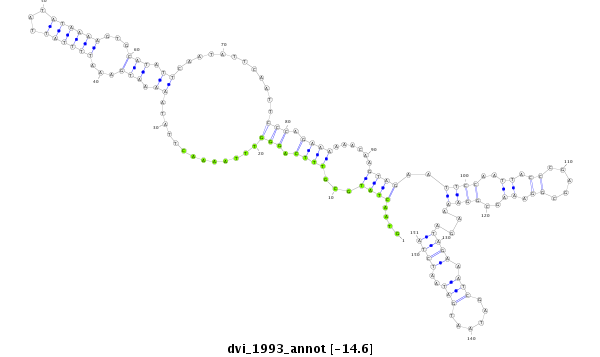
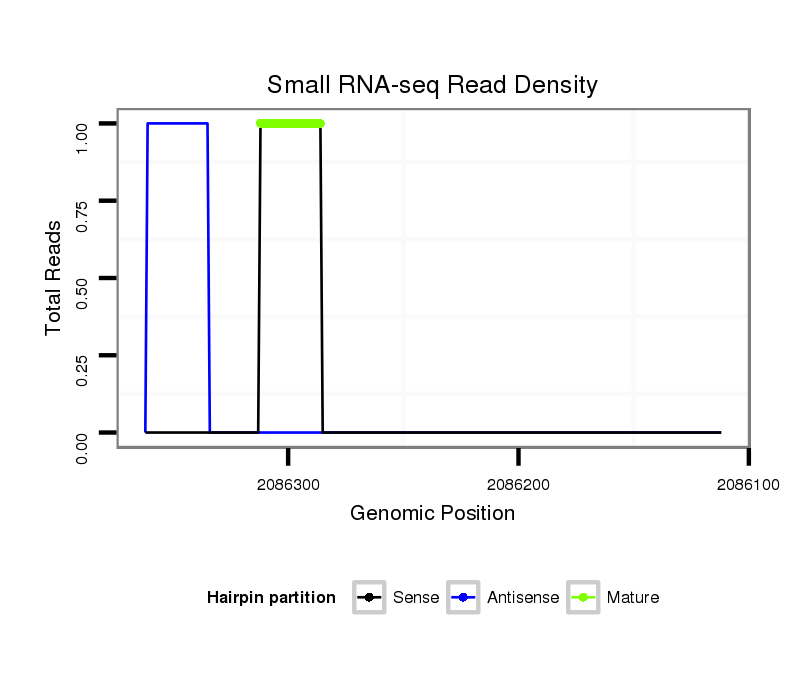
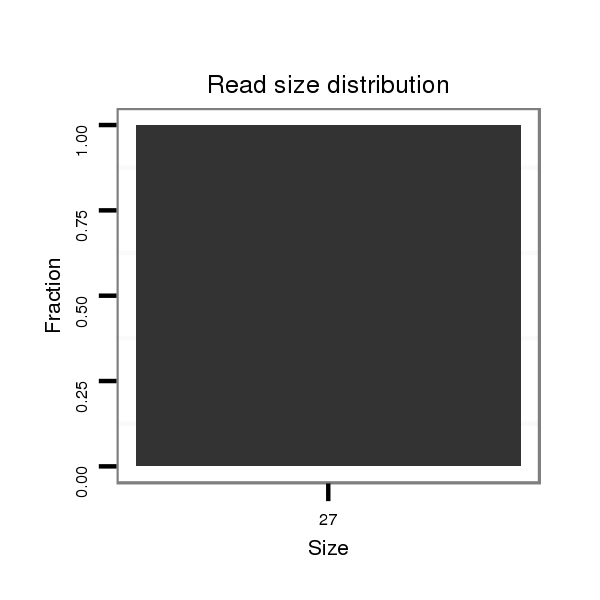
ID:dvi_1993 |
Coordinate:scaffold_12726:2086162-2086312 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -14.5 | -14.5 | -14.2 |
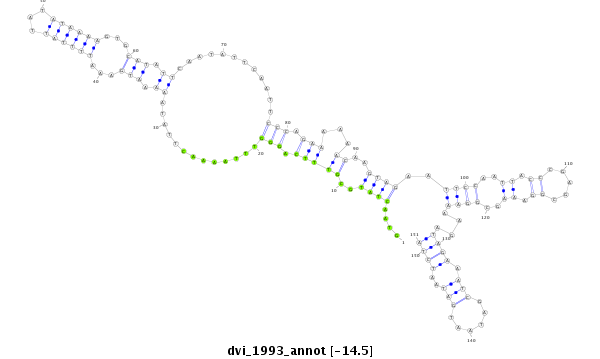 |
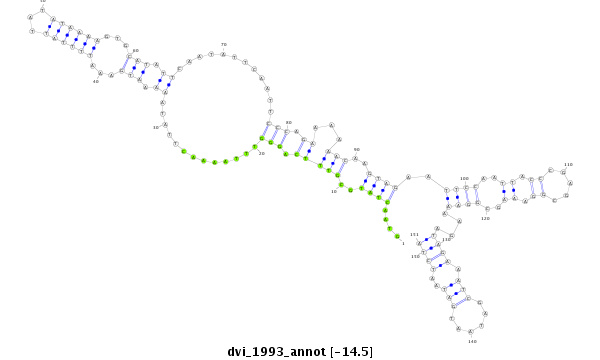 |
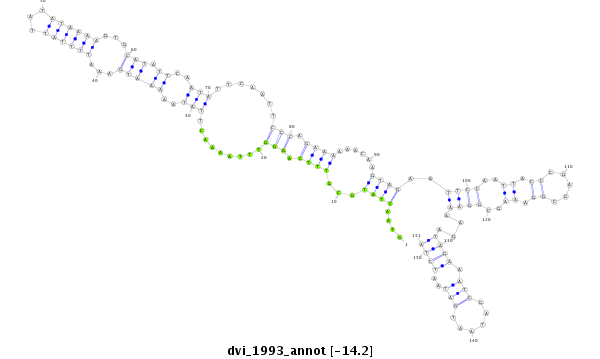 |
exon [dvir_GLEANR_15525:5]; CDS [Dvir\GJ15200-cds]; intron [Dvir\GJ15200-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCAAACTAAATGATTTGCCAGATATTAATGATATAGGATTGTCACAAATGGTAACTATGCGTTTCAGGGTTTAAAACTTATAAAAATGAAATTTTATTATATAAAAGTGCATATTCAATATTCAATTCCCAGAAAAAACAAGTAGAATTCCAATTACCCGAGCGGAAAGCGGAAAAGATAGAAATCGATAATGATAATCTAAAGTTTCTGATTAACAAATACAACGTTTTTAATAATCCACAACAGCCAGT **************************************************....((((...((((.(((.............((.(((...((((((...))))))...))).))............))).))))......))))..((((..((.((.....)).))..))))....((((.(((......)))..))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
V116 male body |
SRR060668 160x9_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
V053 head |
SRR060687 9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAACTATGCGTTTCAGGGTTTAAAAC.............................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GACTTGCCAGAAGTTAATGAT........................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATCAGTATTCAATTGCCAGA...................................................................................................................... | 20 | 3 | 18 | 0.39 | 7 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................TCAGTATTCAATTCTCAGA...................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATCAGTATTCAATTCTCAGA...................................................................................................................... | 20 | 3 | 15 | 0.13 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................TGGTATCGGACTGTCACAA............................................................................................................................................................................................................ | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................TCAGTATTCAATTGCCAGAG..................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GCGGAATCGATAGAAATCGC............................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................AGAGACATCGATAATGATGA...................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................CAGTATTCAATTGCCAGAG..................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATCAGTATTCAATTCACAGA...................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CGTTTGATTTACTAAACGGTCTATAATTACTATATCCTAACAGTGTTTACCATTGATACGCAAAGTCCCAAATTTTGAATATTTTTACTTTAAAATAATATATTTTCACGTATAAGTTATAAGTTAAGGGTCTTTTTTGTTCATCTTAAGGTTAATGGGCTCGCCTTTCGCCTTTTCTATCTTTAGCTATTACTATTAGATTTCAAAGACTAATTGTTTATGTTGCAAAAATTATTAGGTGTTGTCGGTCA
**************************************************....((((...((((.(((.............((.(((...((((((...))))))...))).))............))).))))......))))..((((..((.((.....)).))..))))....((((.(((......)))..))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060667 160_females_carcasses_total |
SRR060675 140x9_ovaries_total |
V053 head |
V116 male body |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TAAGGTTAATGTCCTAGCCTT.................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .GTTTGATTTACTAAACGGTCTATAATT............................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGTTTGATTTGCTAGACGGT....................................................................................................................................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................TGTTGCAGAATTTATTAGG............ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................ATATCGTAACAGTGATTCC......................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| CGTTTGAATTACTATAGGGT....................................................................................................................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| CGTTGGATTTGCTAGACGGT....................................................................................................................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| CGTTCGATTTGCTACACGGT....................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:2086112-2086362 - | dvi_1993 | GCAAACTAAATGATTTGCCAGATATTAATGATATAGGATTGTCACAAATGGTAACTATGCGTTTCAGGGTTTAAAACTTATAAAAATGAAATTTTATTATATAAAAGTGCATATTCAATATTCAATTCCCAGAAAAAACAAGTAGAATTCCAATTACCCGAGCGGAAAGCGGAAAAGATAGAAATCGATAATGATAATCTAAAGTTTCTGATTAACAAATACAACGTTTTTAATAATCCACAACAGCCAGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 04:41 AM