
ID:dvi_19923 |
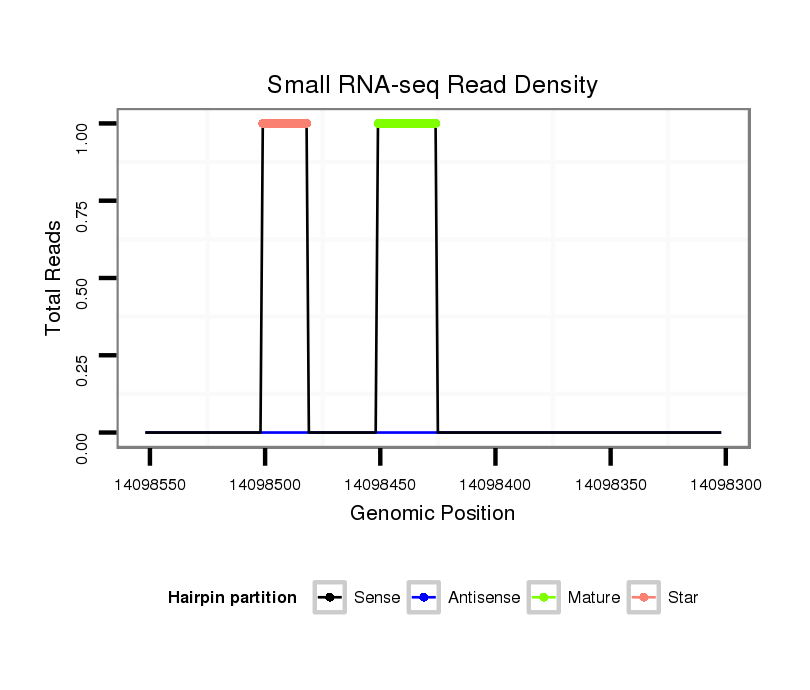
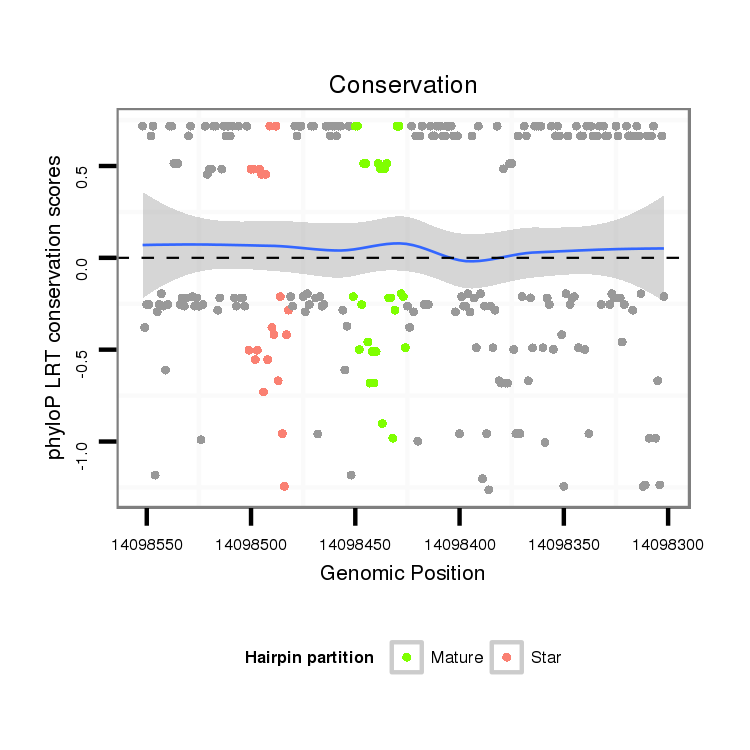
Coordinate:scaffold_13049:14098352-14098502 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -4.3 | -4.3 | -3.9 | -3.6 |
 |
 |
 |
 |
exon [dvir_GLEANR_11820:2]; CDS [Dvir\GJ11807-cds]; intron [Dvir\GJ11807-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTTGTTCACACATTATTATAAGTTTGTATACTATTTAAACACAATACGAATATTCTCAGTTATAGTTCTAATTCTTTCGATTATATTATGAAGTTACCTATAAATTTTCTGTTGCCAGCGTTTCTATTTGGCCTAATTGCATTGGTAAGCGTAAAGCGAGTCGATCACTTGGCCCATATCTAAGGGTTATTATATTTCAGTGCCAATTGCCATATGGCAATGCTGGAGCTTTGGTCAGCCCCCCAGGGGT ***************************************************((....(((((((((......(((...............))).....)))))).....))).............))**************************************************************************************************************************** |
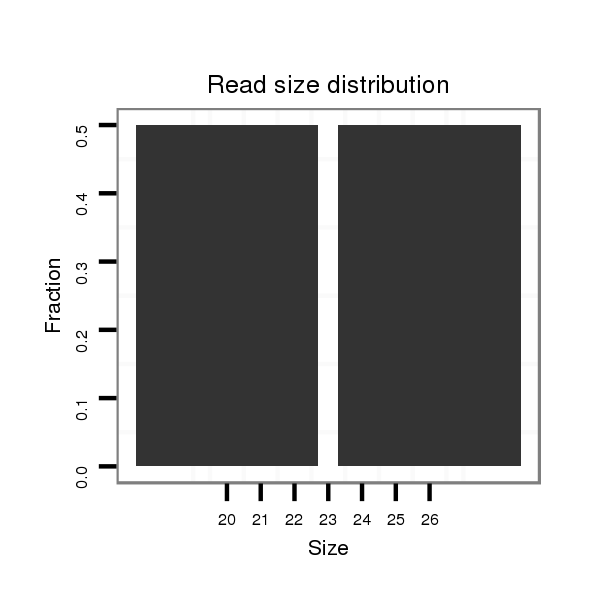
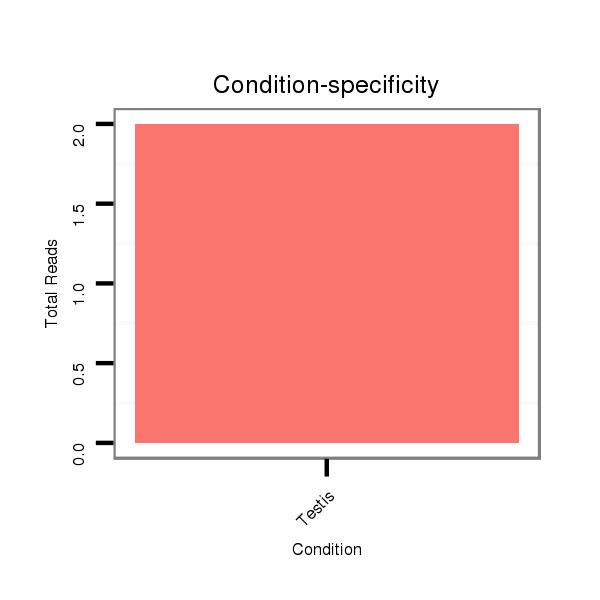
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
SRR060679 140x9_testes_total |
SRR060659 Argentina_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TAAATTTTCTGTTGCCAGCGTTTCTA............................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TATTCTCAGTTATAGTTCTA.................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................TCAAGCGAGGAGATCACTTG............................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................GACAGCGTTTGTATTTGG....................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................ATATTATCAGTTATGGTTC...................................................................................................................................................................................... | 19 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................TAGGGGTTATTATCTATCA................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
AAAACAAGTGTGTAATAATATTCAAACATATGATAAATTTGTGTTATGCTTATAAGAGTCAATATCAAGATTAAGAAAGCTAATATAATACTTCAATGGATATTTAAAAGACAACGGTCGCAAAGATAAACCGGATTAACGTAACCATTCGCATTTCGCTCAGCTAGTGAACCGGGTATAGATTCCCAATAATATAAAGTCACGGTTAACGGTATACCGTTACGACCTCGAAACCAGTCGGGGGGTCCCCA
****************************************************************************************************************************((....(((((((((......(((...............))).....)))))).....))).............))*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060669 160x9_females_carcasses_total |
V047 embryo |
SRR060685 9xArg_0-2h_embryos_total |
M027 male body |
SRR060665 9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................AGTGAACCGAGTACAGATTC.................................................................. | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TCGGATTCCGCCCAGCTAG.................................................................................... | 19 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................ATATTCAAACATTTAAGAAATTT................................................................................................................................................................................................................... | 23 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................CCAGGCGGGGCGTCCCCC | 18 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................ATATTCAAACATTTAAGAAATT.................................................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................TACCGTTACTACCTAGATA.................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:14098302-14098552 - | dvi_19923 | TTTTGTTCACAC------ATTATTATAAGTTTGTATACTATTTAAACACAA--------------TACGAATATTCTCAGTTATA--GT--------------TCTAATTCTTTCGATTATATTATGAAGTTACCTATAAATTTTCTGTTGCCAGCGTTTCTATTTGGCCTAATTGCATTGGTAAGCGTAAAGCGA--GTCGATCACTTGGCCCATATCTAAG-GGTT---AT-------TATATTTCAGTGCCAATTGCCATATGGC-AATGCTGGAGCTTTGGTCAGCCCCCCAGGGGT |
| droMoj3 | scaffold_6680:13208102-13208366 + | TTCCGTGCGCTGTTTATTGTT---GTTAGTTAAATCACTATTCAAACACAA--------------AATATACATATTCCGATAAATATTTGTATTATTTAAGGCAAGAATCTTTCGGTTAAAACATGAAGTTGGTTCTAACC------------ATACTTCTCATTGGACTGGCTGCATTGGTAAGCGCATTGCGATAGTCATCGTCCTT------CCCCAAGTTAGTCTTATTCGTGTATCGACTGCAGATCCGATTGCCCTGTGGCCAGTGCCGAAGT-CTGGCCAGCAGCTCAATCGT | |
| droGri2 | scaffold_15110:16106546-16106817 + | TGTTGTCAATAC-T-GATATTATTAAATGTCTGTCTA---TTCT-ACACATAACTTACAACGCACTACGAA----------TCAAAGTCTAGATTATTAATAGAGAGGTTCTTCAAATTTCGTTATGAAGTTGCTTGCAAATTTCAATATGGCAGCTCTTTCCTGTTGTCTAATTGCATTGGTAAGAGAGAAAAGT--CTTCAAAAGCTTTCAAATAACAGAGATGGT---AT-------TCATTTTCAGACTCATCTCCCATTTGGC-AATGCTAGAGCCTTGGCCAGTGTCACATTAGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 04:49 AM